Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 9610-11-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2283 | 1.915e-02 | 1.563e-02 | 1.445e-01 |
|---|
| IPC-NIBC-129 | 0.1942 | 4.594e-02 | 6.636e-02 | 4.217e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2182 | 4.804e-03 | 8.513e-03 | 1.787e-01 |
|---|
| Loi_GPL570 | 0.2970 | 2.988e-02 | 3.445e-02 | 1.433e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1785 | 1.560e-04 | 8.410e-04 | 3.514e-03 |
|---|
| Parker_GPL1390 | 0.2735 | 5.997e-02 | 7.386e-02 | 4.344e-01 |
|---|
| Parker_GPL887 | 0.1598 | 3.018e-01 | 3.198e-01 | 3.206e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.2910 | 3.093e-01 | 2.395e-01 | 8.745e-01 |
|---|
| Schmidt | 0.2303 | 1.603e-02 | 1.826e-02 | 2.009e-01 |
|---|
| Sotiriou | 0.3511 | 6.327e-03 | 4.776e-03 | 1.443e-01 |
|---|
| Van-De-Vijver | 0.2631 | 3.227e-03 | 4.628e-03 | 1.251e-01 |
|---|
| Zhang | 0.2841 | 1.015e-01 | 6.192e-02 | 2.222e-01 |
|---|
| Zhou | 0.3546 | 1.239e-01 | 1.217e-01 | 5.920e-01 |
|---|
Expression data for subnetwork 9610-11-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
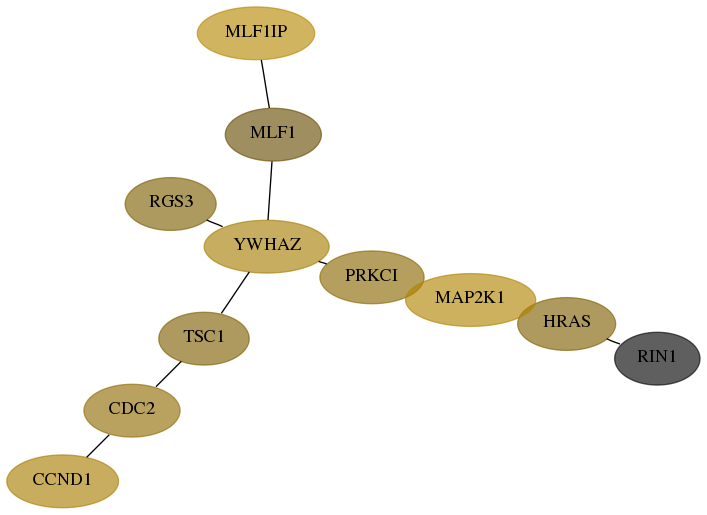
Score for each gene in subnetwork 9610-11-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| mlf1 |   | 7 | 30 | 56 | 49 | 0.027 | 0.224 | 0.053 | 0.047 | 0.169 | 0.128 | 0.021 | -0.028 | 0.106 | 0.176 | undef | 0.098 | 0.141 |
|---|
| rgs3 |   | 4 | 50 | 77 | 66 | 0.048 | 0.195 | 0.029 | 0.052 | 0.056 | 0.192 | -0.052 | -0.078 | 0.033 | 0.023 | undef | 0.100 | -0.196 |
|---|
| mlf1ip |   | 5 | 40 | 77 | 64 | 0.182 | 0.147 | 0.186 | 0.144 | 0.242 | 0.266 | 0.005 | 0.345 | 0.182 | 0.197 | undef | 0.240 | 0.372 |
|---|
| ywhaz |   | 17 | 10 | 29 | 25 | 0.124 | 0.089 | 0.087 | 0.200 | 0.141 | 0.110 | 0.228 | 0.199 | 0.056 | 0.123 | undef | 0.103 | 0.213 |
|---|
| prkci |   | 14 | 15 | 31 | 26 | 0.065 | 0.104 | 0.012 | 0.170 | 0.112 | 0.088 | -0.103 | -0.170 | 0.196 | 0.128 | 0.049 | 0.124 | 0.149 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| hras |   | 14 | 15 | 42 | 34 | 0.049 | -0.030 | 0.094 | 0.210 | 0.160 | 0.195 | 0.022 | 0.245 | 0.086 | 0.208 | 0.180 | 0.003 | 0.197 |
|---|
| rin1 |   | 1 | 116 | 162 | 169 | -0.000 | 0.007 | -0.017 | 0.073 | 0.074 | -0.083 | -0.239 | 0.089 | 0.097 | -0.025 | 0.109 | 0.072 | -0.195 |
|---|
| map2k1 |   | 10 | 20 | 54 | 44 | 0.154 | 0.096 | 0.093 | 0.013 | 0.095 | 0.045 | -0.223 | 0.207 | 0.082 | 0.096 | undef | undef | 0.230 |
|---|
GO Enrichment output for subnetwork 9610-11-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.492E-08 | 3.431E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.45E-07 | 2.817E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 8.282E-07 | 6.349E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.681E-06 | 9.664E-04 |
|---|
| sperm motility | GO:0030317 |  | 2.975E-06 | 1.368E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 5.35E-06 | 2.051E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 7.245E-06 | 2.381E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 8.728E-06 | 2.509E-03 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 1.227E-05 | 3.135E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.371E-05 | 3.154E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.371E-05 | 2.867E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 4.373E-09 | 1.068E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.245E-07 | 1.521E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.435E-07 | 1.983E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.124E-06 | 6.868E-04 |
|---|
| sperm motility | GO:0030317 |  | 1.415E-06 | 6.915E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 2.138E-06 | 8.706E-04 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 3.072E-06 | 1.072E-03 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 4.923E-06 | 1.503E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.975E-06 | 1.622E-03 |
|---|
| DNA replication. synthesis of RNA primer | GO:0006269 |  | 5.975E-06 | 1.46E-03 |
|---|
| hemopoietic progenitor cell differentiation | GO:0002244 |  | 5.975E-06 | 1.327E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 7.187E-09 | 1.729E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.045E-07 | 2.461E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.997E-07 | 3.206E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.438E-06 | 8.647E-04 |
|---|
| sperm motility | GO:0030317 |  | 1.633E-06 | 7.856E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 3.505E-06 | 1.406E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 5.034E-06 | 1.73E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 7.492E-06 | 2.253E-03 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 8.063E-06 | 2.156E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.321E-06 | 2.002E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 8.321E-06 | 1.82E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 4.373E-09 | 1.068E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.245E-07 | 1.521E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.435E-07 | 1.983E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.124E-06 | 6.868E-04 |
|---|
| sperm motility | GO:0030317 |  | 1.415E-06 | 6.915E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 2.138E-06 | 8.706E-04 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 3.072E-06 | 1.072E-03 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 4.923E-06 | 1.503E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.975E-06 | 1.622E-03 |
|---|
| DNA replication. synthesis of RNA primer | GO:0006269 |  | 5.975E-06 | 1.46E-03 |
|---|
| hemopoietic progenitor cell differentiation | GO:0002244 |  | 5.975E-06 | 1.327E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 7.187E-09 | 1.729E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.045E-07 | 2.461E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.997E-07 | 3.206E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.438E-06 | 8.647E-04 |
|---|
| sperm motility | GO:0030317 |  | 1.633E-06 | 7.856E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 3.505E-06 | 1.406E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 5.034E-06 | 1.73E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 7.492E-06 | 2.253E-03 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 8.063E-06 | 2.156E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.321E-06 | 2.002E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 8.321E-06 | 1.82E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 4.142E-06 | 0.01478934 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.21E-06 | 0.01108799 |
|---|
| hemopoietic progenitor cell differentiation | GO:0002244 |  | 6.21E-06 | 7.392E-03 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 6.21E-06 | 5.544E-03 |
|---|
| establishment of apical/basal cell polarity | GO:0035089 |  | 6.21E-06 | 4.435E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 8.691E-06 | 5.173E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 8.691E-06 | 4.434E-03 |
|---|
| eye photoreceptor cell differentiation | GO:0001754 |  | 8.691E-06 | 3.879E-03 |
|---|
| regulation of synaptic transmission. GABAergic | GO:0032228 |  | 8.691E-06 | 3.448E-03 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.158E-05 | 4.137E-03 |
|---|
| positive regulation of small GTPase mediated signal transduction | GO:0051057 |  | 1.158E-05 | 3.76E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| hemopoietic progenitor cell differentiation | GO:0002244 |  | 3.136E-06 | 0.0114404 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 3.136E-06 | 5.72E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 3.136E-06 | 3.813E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.703E-06 | 4.289E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 4.703E-06 | 3.431E-03 |
|---|
| regulation of synaptic transmission. GABAergic | GO:0032228 |  | 4.703E-06 | 2.859E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 4.703E-06 | 2.451E-03 |
|---|
| regulation of long-term neuronal synaptic plasticity | GO:0048169 |  | 6.582E-06 | 3.001E-03 |
|---|
| establishment or maintenance of epithelial cell apical/basal polarity | GO:0045197 |  | 6.582E-06 | 2.668E-03 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 6.582E-06 | 2.401E-03 |
|---|
| cell cortex | GO:0005938 |  | 8.086E-06 | 2.682E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 7.187E-09 | 1.729E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.045E-07 | 2.461E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.997E-07 | 3.206E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.438E-06 | 8.647E-04 |
|---|
| sperm motility | GO:0030317 |  | 1.633E-06 | 7.856E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 3.505E-06 | 1.406E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 5.034E-06 | 1.73E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 7.492E-06 | 2.253E-03 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 8.063E-06 | 2.156E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.321E-06 | 2.002E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 8.321E-06 | 1.82E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.492E-08 | 3.431E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.45E-07 | 2.817E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 8.282E-07 | 6.349E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.681E-06 | 9.664E-04 |
|---|
| sperm motility | GO:0030317 |  | 2.975E-06 | 1.368E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 5.35E-06 | 2.051E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 7.245E-06 | 2.381E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 8.728E-06 | 2.509E-03 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 1.227E-05 | 3.135E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.371E-05 | 3.154E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.371E-05 | 2.867E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.492E-08 | 3.431E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.45E-07 | 2.817E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 8.282E-07 | 6.349E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.681E-06 | 9.664E-04 |
|---|
| sperm motility | GO:0030317 |  | 2.975E-06 | 1.368E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 5.35E-06 | 2.051E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 7.245E-06 | 2.381E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 8.728E-06 | 2.509E-03 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 1.227E-05 | 3.135E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.371E-05 | 3.154E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.371E-05 | 2.867E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of long-term neuronal synaptic plasticity | GO:0048169 |  | 4.222E-06 | 7.781E-03 |
|---|
| establishment or maintenance of epithelial cell apical/basal polarity | GO:0045197 |  | 4.222E-06 | 3.891E-03 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 4.222E-06 | 2.594E-03 |
|---|
| establishment of cell polarity | GO:0030010 |  | 4.222E-06 | 1.945E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 6.331E-06 | 2.334E-03 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 6.331E-06 | 1.945E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 6.331E-06 | 1.667E-03 |
|---|
| establishment or maintenance of apical/basal cell polarity | GO:0035088 |  | 6.331E-06 | 1.459E-03 |
|---|
| regulation of synaptic transmission. GABAergic | GO:0032228 |  | 6.331E-06 | 1.297E-03 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 8.861E-06 | 1.633E-03 |
|---|
| visual behavior | GO:0007632 |  | 8.861E-06 | 1.485E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.492E-08 | 3.431E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.45E-07 | 2.817E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 8.282E-07 | 6.349E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.681E-06 | 9.664E-04 |
|---|
| sperm motility | GO:0030317 |  | 2.975E-06 | 1.368E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 5.35E-06 | 2.051E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 7.245E-06 | 2.381E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 8.728E-06 | 2.509E-03 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 1.227E-05 | 3.135E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.371E-05 | 3.154E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.371E-05 | 2.867E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.173E-08 | 2.784E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.568E-07 | 3.047E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 7.909E-07 | 6.256E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.785E-06 | 1.059E-03 |
|---|
| sperm motility | GO:0030317 |  | 3.005E-06 | 1.426E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 5.176E-06 | 2.047E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 6.876E-06 | 2.331E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 8.908E-06 | 2.642E-03 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 1.13E-05 | 2.98E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.166E-05 | 2.766E-03 |
|---|
| establishment of apical/basal cell polarity | GO:0035089 |  | 1.166E-05 | 2.515E-03 |
|---|



