Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 9232-11-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2029 | 1.800e-02 | 1.457e-02 | 1.379e-01 |
|---|
| IPC-NIBC-129 | 0.2960 | 3.173e-02 | 4.654e-02 | 3.282e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2212 | 6.634e-03 | 1.144e-02 | 2.161e-01 |
|---|
| Loi_GPL570 | 0.2011 | 1.156e-01 | 1.284e-01 | 4.086e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1662 | 0.000e+00 | 2.000e-06 | 0.000e+00 |
|---|
| Parker_GPL1390 | 0.2587 | 5.890e-02 | 7.260e-02 | 4.303e-01 |
|---|
| Parker_GPL887 | 0.2055 | 3.533e-01 | 3.720e-01 | 3.842e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3919 | 1.944e-01 | 1.373e-01 | 7.437e-01 |
|---|
| Schmidt | 0.2315 | 2.941e-02 | 3.487e-02 | 3.015e-01 |
|---|
| Sotiriou | 0.3206 | 1.054e-02 | 8.073e-03 | 1.941e-01 |
|---|
| Van-De-Vijver | 0.2900 | 1.542e-03 | 2.217e-03 | 7.815e-02 |
|---|
| Zhang | 0.2472 | 1.223e-01 | 7.830e-02 | 2.599e-01 |
|---|
| Zhou | 0.3961 | 4.957e-02 | 3.736e-02 | 3.126e-01 |
|---|
Expression data for subnetwork 9232-11-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
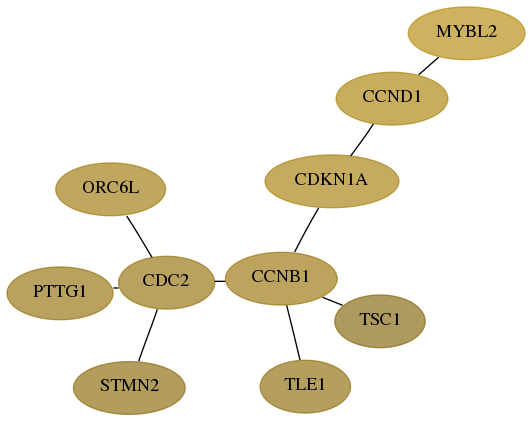
Score for each gene in subnetwork 9232-11-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| mybl2 |   | 9 | 22 | 56 | 48 | 0.162 | 0.237 | 0.166 | 0.079 | 0.124 | 0.184 | 0.333 | 0.275 | 0.202 | 0.182 | 0.289 | 0.074 | 0.245 |
|---|
| stmn2 |   | 29 | 9 | 34 | 27 | 0.057 | 0.106 | 0.005 | 0.052 | 0.110 | 0.071 | -0.099 | 0.006 | 0.034 | 0.103 | 0.008 | 0.113 | -0.039 |
|---|
| cdkn1a |   | 44 | 7 | 1 | 4 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | undef | -0.201 | 0.069 |
|---|
| tle1 |   | 47 | 6 | 1 | 3 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | 0.170 | 0.068 | 0.025 |
|---|
| pttg1 |   | 1 | 116 | 111 | 122 | 0.072 | 0.270 | 0.175 | 0.183 | 0.200 | 0.208 | 0.347 | 0.334 | 0.224 | 0.204 | 0.200 | 0.210 | 0.309 |
|---|
| ccnb1 |   | 42 | 8 | 56 | 39 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | undef | 0.167 | 0.398 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| orc6l |   | 15 | 11 | 34 | 28 | 0.094 | 0.270 | 0.084 | 0.114 | 0.106 | 0.240 | 0.256 | 0.234 | 0.192 | 0.105 | 0.258 | 0.175 | 0.160 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
GO Enrichment output for subnetwork 9232-11-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 8.282E-07 | 1.905E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 9.496E-07 | 1.092E-03 |
|---|
| interphase | GO:0051325 |  | 1.042E-06 | 7.986E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.591E-06 | 4.365E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.841E-06 | 3.607E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 7.841E-06 | 3.006E-03 |
|---|
| response to UV | GO:0009411 |  | 9.322E-06 | 3.063E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.275E-05 | 3.666E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.645E-05 | 4.203E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.645E-05 | 3.783E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.192E-05 | 4.582E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.04E-07 | 4.983E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.956E-07 | 3.611E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.937E-06 | 1.578E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.705E-06 | 1.652E-03 |
|---|
| response to UV | GO:0009411 |  | 3.338E-06 | 1.631E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.72E-06 | 1.515E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 4.055E-06 | 1.415E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 6.348E-06 | 1.939E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 6.635E-06 | 1.801E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 7.545E-06 | 1.843E-03 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 7.545E-06 | 1.676E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.472E-07 | 8.354E-04 |
|---|
| interphase | GO:0051325 |  | 3.935E-07 | 4.734E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.853E-07 | 3.892E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.176E-06 | 1.911E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.766E-06 | 1.812E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 3.766E-06 | 1.51E-03 |
|---|
| response to UV | GO:0009411 |  | 4.888E-06 | 1.68E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.096E-06 | 1.833E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.039E-05 | 2.779E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.053E-05 | 2.534E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.053E-05 | 2.304E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.04E-07 | 4.983E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.956E-07 | 3.611E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.937E-06 | 1.578E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.705E-06 | 1.652E-03 |
|---|
| response to UV | GO:0009411 |  | 3.338E-06 | 1.631E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.72E-06 | 1.515E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 4.055E-06 | 1.415E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 6.348E-06 | 1.939E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 6.635E-06 | 1.801E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 7.545E-06 | 1.843E-03 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 7.545E-06 | 1.676E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.472E-07 | 8.354E-04 |
|---|
| interphase | GO:0051325 |  | 3.935E-07 | 4.734E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.853E-07 | 3.892E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.176E-06 | 1.911E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.766E-06 | 1.812E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 3.766E-06 | 1.51E-03 |
|---|
| response to UV | GO:0009411 |  | 4.888E-06 | 1.68E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.096E-06 | 1.833E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.039E-05 | 2.779E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.053E-05 | 2.534E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.053E-05 | 2.304E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.259E-08 | 4.494E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 3.704E-08 | 6.614E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.523E-07 | 1.813E-04 |
|---|
| regulation of Ras GTPase activity | GO:0032318 |  | 1.651E-07 | 1.474E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.824E-07 | 1.303E-04 |
|---|
| regulation of GTPase activity | GO:0043087 |  | 3.596E-07 | 2.14E-04 |
|---|
| cysteine-type endopeptidase inhibitor activity | GO:0004869 |  | 9.053E-07 | 4.618E-04 |
|---|
| regulation of Rab protein signal transduction | GO:0032483 |  | 2.195E-06 | 9.799E-04 |
|---|
| Rab GTPase activator activity | GO:0005097 |  | 2.368E-06 | 9.395E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.366E-06 | 1.202E-03 |
|---|
| response to UV | GO:0009411 |  | 4.081E-06 | 1.325E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.125E-08 | 4.104E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 7.296E-08 | 1.331E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.122E-07 | 1.364E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.568E-07 | 1.43E-04 |
|---|
| interphase | GO:0051325 |  | 1.731E-07 | 1.263E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.448E-06 | 1.488E-03 |
|---|
| response to UV | GO:0009411 |  | 2.813E-06 | 1.466E-03 |
|---|
| origin recognition complex | GO:0000808 |  | 3.763E-06 | 1.716E-03 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 3.763E-06 | 1.525E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 3.763E-06 | 1.373E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.119E-06 | 1.366E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.472E-07 | 8.354E-04 |
|---|
| interphase | GO:0051325 |  | 3.935E-07 | 4.734E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.853E-07 | 3.892E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.176E-06 | 1.911E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.766E-06 | 1.812E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 3.766E-06 | 1.51E-03 |
|---|
| response to UV | GO:0009411 |  | 4.888E-06 | 1.68E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.096E-06 | 1.833E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.039E-05 | 2.779E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.053E-05 | 2.534E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.053E-05 | 2.304E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 8.282E-07 | 1.905E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 9.496E-07 | 1.092E-03 |
|---|
| interphase | GO:0051325 |  | 1.042E-06 | 7.986E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.591E-06 | 4.365E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.841E-06 | 3.607E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 7.841E-06 | 3.006E-03 |
|---|
| response to UV | GO:0009411 |  | 9.322E-06 | 3.063E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.275E-05 | 3.666E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.645E-05 | 4.203E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.645E-05 | 3.783E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.192E-05 | 4.582E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 8.282E-07 | 1.905E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 9.496E-07 | 1.092E-03 |
|---|
| interphase | GO:0051325 |  | 1.042E-06 | 7.986E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.591E-06 | 4.365E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.841E-06 | 3.607E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 7.841E-06 | 3.006E-03 |
|---|
| response to UV | GO:0009411 |  | 9.322E-06 | 3.063E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.275E-05 | 3.666E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.645E-05 | 4.203E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.645E-05 | 3.783E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.192E-05 | 4.582E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.329E-05 | 0.02448556 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 1.329E-05 | 0.01224278 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 1.329E-05 | 8.162E-03 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 1.859E-05 | 8.566E-03 |
|---|
| regulation of stress fiber formation | GO:0051492 |  | 2.478E-05 | 9.132E-03 |
|---|
| regulation of dephosphorylation | GO:0035303 |  | 3.184E-05 | 9.78E-03 |
|---|
| regulation of actin filament bundle formation | GO:0032231 |  | 3.184E-05 | 8.383E-03 |
|---|
| negative regulation of signal transduction | GO:0009968 |  | 3.579E-05 | 8.245E-03 |
|---|
| regulation of protein ubiquitination | GO:0031396 |  | 3.978E-05 | 8.146E-03 |
|---|
| negative regulation of cell communication | GO:0010648 |  | 4.393E-05 | 8.096E-03 |
|---|
| negative regulation of translation | GO:0017148 |  | 5.829E-05 | 9.766E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 8.282E-07 | 1.905E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 9.496E-07 | 1.092E-03 |
|---|
| interphase | GO:0051325 |  | 1.042E-06 | 7.986E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.591E-06 | 4.365E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.841E-06 | 3.607E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 7.841E-06 | 3.006E-03 |
|---|
| response to UV | GO:0009411 |  | 9.322E-06 | 3.063E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.275E-05 | 3.666E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.645E-05 | 4.203E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.645E-05 | 3.783E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.192E-05 | 4.582E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 6.834E-07 | 1.622E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.784E-07 | 9.235E-04 |
|---|
| interphase | GO:0051325 |  | 8.529E-07 | 6.747E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.723E-06 | 3.988E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.867E-06 | 3.259E-03 |
|---|
| response to UV | GO:0009411 |  | 8.218E-06 | 3.25E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.029E-05 | 3.49E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.184E-05 | 3.511E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.441E-05 | 3.798E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 1.92E-05 | 4.556E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.92E-05 | 4.142E-03 |
|---|



