Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 875-10-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2119 | 6.967e-03 | 4.967e-03 | 6.517e-02 |
|---|
| IPC-NIBC-129 | 0.2395 | 3.174e-02 | 4.655e-02 | 3.283e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2674 | 2.310e-03 | 4.352e-03 | 1.134e-01 |
|---|
| Loi_GPL570 | 0.1333 | 2.830e-01 | 3.050e-01 | 7.049e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1676 | 6.000e-06 | 5.700e-05 | 2.700e-05 |
|---|
| Parker_GPL1390 | 0.3069 | 1.231e-01 | 1.462e-01 | 6.162e-01 |
|---|
| Parker_GPL887 | 0.2045 | 3.343e-01 | 3.528e-01 | 3.607e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3803 | 1.963e-01 | 1.390e-01 | 7.468e-01 |
|---|
| Schmidt | 0.1792 | 2.638e-02 | 3.107e-02 | 2.811e-01 |
|---|
| Sotiriou | 0.2668 | 2.567e-02 | 2.022e-02 | 3.157e-01 |
|---|
| Van-De-Vijver | 0.2900 | 8.600e-04 | 1.238e-03 | 5.304e-02 |
|---|
| Zhang | 0.2538 | 1.198e-01 | 7.629e-02 | 2.554e-01 |
|---|
| Zhou | 0.4288 | 5.518e-02 | 4.302e-02 | 3.401e-01 |
|---|
Expression data for subnetwork 875-10-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
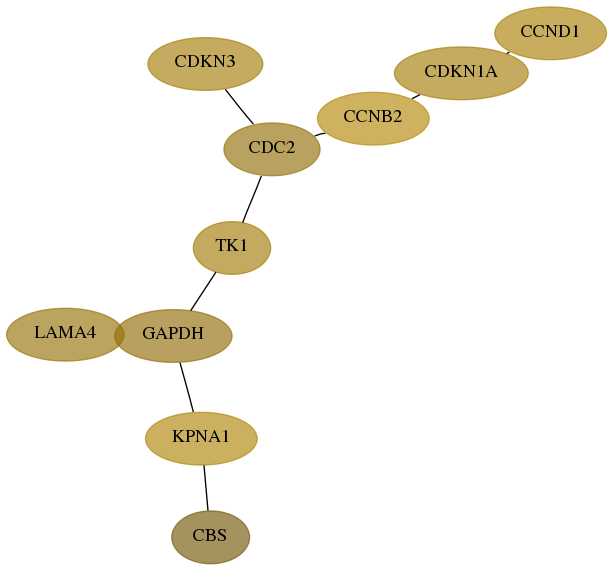
Score for each gene in subnetwork 875-10-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| cdkn3 |   | 53 | 5 | 29 | 22 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.166 | -0.009 | 0.323 | 0.186 | 0.198 | 0.201 | 0.271 | 0.372 |
|---|
| ccnb2 |   | 12 | 18 | 84 | 62 | 0.164 | 0.176 | 0.204 | 0.123 | 0.172 | 0.178 | 0.090 | 0.366 | 0.240 | 0.169 | 0.279 | 0.211 | 0.463 |
|---|
| cdkn1a |   | 44 | 7 | 1 | 4 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | undef | -0.201 | 0.069 |
|---|
| kpna1 |   | 2 | 78 | 153 | 146 | 0.147 | 0.079 | 0.132 | -0.037 | 0.109 | 0.198 | -0.559 | 0.256 | 0.098 | 0.096 | 0.109 | 0.147 | 0.107 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| lama4 |   | 6 | 35 | 84 | 73 | 0.079 | 0.190 | 0.023 | -0.039 | 0.041 | 0.126 | -0.167 | -0.168 | -0.007 | -0.089 | 0.028 | 0.057 | 0.089 |
|---|
| gapdh |   | 6 | 35 | 18 | 20 | 0.071 | 0.312 | 0.102 | 0.145 | 0.165 | 0.172 | -0.253 | 0.288 | 0.064 | 0.213 | 0.225 | -0.088 | 0.235 |
|---|
| tk1 |   | 7 | 30 | 84 | 67 | 0.109 | 0.149 | 0.190 | -0.017 | 0.131 | 0.105 | -0.100 | 0.257 | 0.204 | 0.142 | 0.293 | 0.164 | 0.222 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| cbs |   | 1 | 116 | 153 | 158 | 0.034 | 0.162 | 0.074 | -0.095 | 0.113 | 0.266 | 0.067 | 0.125 | -0.055 | 0.049 | 0.034 | 0.140 | 0.213 |
|---|
GO Enrichment output for subnetwork 875-10-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.715E-07 | 6.245E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.46E-07 | 6.279E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.5E-06 | 3.45E-03 |
|---|
| interphase | GO:0051325 |  | 4.933E-06 | 2.837E-03 |
|---|
| cysteine metabolic process | GO:0006534 |  | 2.476E-05 | 0.01138808 |
|---|
| glycolysis | GO:0006096 |  | 2.543E-05 | 9.75E-03 |
|---|
| response to UV | GO:0009411 |  | 2.91E-05 | 9.562E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.463E-05 | 9.957E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 4.614E-05 | 0.01179095 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 4.614E-05 | 0.01061186 |
|---|
| regulation of embryonic development | GO:0045995 |  | 4.614E-05 | 9.647E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.527E-08 | 1.839E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.81E-07 | 2.211E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.614E-06 | 1.315E-03 |
|---|
| glycolysis | GO:0006096 |  | 8.889E-06 | 5.429E-03 |
|---|
| response to UV | GO:0009411 |  | 1.528E-05 | 7.466E-03 |
|---|
| cysteine metabolic process | GO:0006534 |  | 1.542E-05 | 6.277E-03 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.614E-05 | 5.632E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.054E-05 | 6.274E-03 |
|---|
| hexose catabolic process | GO:0019320 |  | 2.897E-05 | 7.863E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.298E-05 | 8.058E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 3.298E-05 | 7.325E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.884E-08 | 1.416E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.415E-07 | 1.703E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.117E-06 | 8.957E-04 |
|---|
| interphase | GO:0051325 |  | 1.265E-06 | 7.61E-04 |
|---|
| glycolysis | GO:0006096 |  | 7.006E-06 | 3.371E-03 |
|---|
| cysteine metabolic process | GO:0006534 |  | 9.903E-06 | 3.971E-03 |
|---|
| response to UV | GO:0009411 |  | 1.152E-05 | 3.958E-03 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.288E-05 | 3.874E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.847E-05 | 4.937E-03 |
|---|
| hexose catabolic process | GO:0019320 |  | 2.337E-05 | 5.623E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.965E-05 | 6.485E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.527E-08 | 1.839E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.81E-07 | 2.211E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.614E-06 | 1.315E-03 |
|---|
| glycolysis | GO:0006096 |  | 8.889E-06 | 5.429E-03 |
|---|
| response to UV | GO:0009411 |  | 1.528E-05 | 7.466E-03 |
|---|
| cysteine metabolic process | GO:0006534 |  | 1.542E-05 | 6.277E-03 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.614E-05 | 5.632E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.054E-05 | 6.274E-03 |
|---|
| hexose catabolic process | GO:0019320 |  | 2.897E-05 | 7.863E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.298E-05 | 8.058E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 3.298E-05 | 7.325E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.884E-08 | 1.416E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.415E-07 | 1.703E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.117E-06 | 8.957E-04 |
|---|
| interphase | GO:0051325 |  | 1.265E-06 | 7.61E-04 |
|---|
| glycolysis | GO:0006096 |  | 7.006E-06 | 3.371E-03 |
|---|
| cysteine metabolic process | GO:0006534 |  | 9.903E-06 | 3.971E-03 |
|---|
| response to UV | GO:0009411 |  | 1.152E-05 | 3.958E-03 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.288E-05 | 3.874E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.847E-05 | 4.937E-03 |
|---|
| hexose catabolic process | GO:0019320 |  | 2.337E-05 | 5.623E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.965E-05 | 6.485E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 8.812E-09 | 3.147E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.036E-08 | 1.849E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.46E-08 | 1.738E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 2.594E-08 | 2.316E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.099E-07 | 7.85E-05 |
|---|
| response to UV | GO:0009411 |  | 2.863E-06 | 1.704E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.969E-06 | 2.535E-03 |
|---|
| cysteine metabolic process | GO:0006534 |  | 4.969E-06 | 2.218E-03 |
|---|
| nuclear localization sequence binding | GO:0008139 |  | 4.969E-06 | 1.972E-03 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 6.445E-06 | 2.301E-03 |
|---|
| laminin-1 complex | GO:0005606 |  | 6.954E-06 | 2.258E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.869E-09 | 2.871E-05 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 8.439E-09 | 1.539E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.59E-08 | 1.933E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 5.475E-08 | 4.993E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.049E-07 | 7.652E-05 |
|---|
| interphase | GO:0051325 |  | 1.158E-07 | 7.039E-05 |
|---|
| response to UV | GO:0009411 |  | 2.113E-06 | 1.101E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 3.136E-06 | 1.43E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.703E-06 | 1.906E-03 |
|---|
| cysteine metabolic process | GO:0006534 |  | 4.703E-06 | 1.716E-03 |
|---|
| laminin-1 complex | GO:0005606 |  | 6.582E-06 | 2.183E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.884E-08 | 1.416E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.415E-07 | 1.703E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.117E-06 | 8.957E-04 |
|---|
| interphase | GO:0051325 |  | 1.265E-06 | 7.61E-04 |
|---|
| glycolysis | GO:0006096 |  | 7.006E-06 | 3.371E-03 |
|---|
| cysteine metabolic process | GO:0006534 |  | 9.903E-06 | 3.971E-03 |
|---|
| response to UV | GO:0009411 |  | 1.152E-05 | 3.958E-03 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.288E-05 | 3.874E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.847E-05 | 4.937E-03 |
|---|
| hexose catabolic process | GO:0019320 |  | 2.337E-05 | 5.623E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.965E-05 | 6.485E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.715E-07 | 6.245E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.46E-07 | 6.279E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.5E-06 | 3.45E-03 |
|---|
| interphase | GO:0051325 |  | 4.933E-06 | 2.837E-03 |
|---|
| cysteine metabolic process | GO:0006534 |  | 2.476E-05 | 0.01138808 |
|---|
| glycolysis | GO:0006096 |  | 2.543E-05 | 9.75E-03 |
|---|
| response to UV | GO:0009411 |  | 2.91E-05 | 9.562E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.463E-05 | 9.957E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 4.614E-05 | 0.01179095 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 4.614E-05 | 0.01061186 |
|---|
| regulation of embryonic development | GO:0045995 |  | 4.614E-05 | 9.647E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.715E-07 | 6.245E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.46E-07 | 6.279E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.5E-06 | 3.45E-03 |
|---|
| interphase | GO:0051325 |  | 4.933E-06 | 2.837E-03 |
|---|
| cysteine metabolic process | GO:0006534 |  | 2.476E-05 | 0.01138808 |
|---|
| glycolysis | GO:0006096 |  | 2.543E-05 | 9.75E-03 |
|---|
| response to UV | GO:0009411 |  | 2.91E-05 | 9.562E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.463E-05 | 9.957E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 4.614E-05 | 0.01179095 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 4.614E-05 | 0.01061186 |
|---|
| regulation of embryonic development | GO:0045995 |  | 4.614E-05 | 9.647E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| thymus development | GO:0048538 |  | 9.493E-06 | 0.01749647 |
|---|
| regulation of embryonic development | GO:0045995 |  | 9.493E-06 | 8.748E-03 |
|---|
| vascular endothelial growth factor receptor signaling pathway | GO:0048010 |  | 1.329E-05 | 8.162E-03 |
|---|
| NLS-bearing substrate import into nucleus | GO:0006607 |  | 1.329E-05 | 6.121E-03 |
|---|
| T cell homeostasis | GO:0043029 |  | 1.771E-05 | 6.527E-03 |
|---|
| lymphocyte homeostasis | GO:0002260 |  | 2.844E-05 | 8.735E-03 |
|---|
| blood vessel development | GO:0001568 |  | 4.796E-05 | 0.01262677 |
|---|
| regulation of DNA recombination | GO:0000018 |  | 4.923E-05 | 0.01134186 |
|---|
| vasculature development | GO:0001944 |  | 5.109E-05 | 0.01046129 |
|---|
| leukocyte homeostasis | GO:0001776 |  | 6.622E-05 | 0.01220484 |
|---|
| glycolysis | GO:0006096 |  | 2.041E-04 | 0.03419627 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.715E-07 | 6.245E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.46E-07 | 6.279E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.5E-06 | 3.45E-03 |
|---|
| interphase | GO:0051325 |  | 4.933E-06 | 2.837E-03 |
|---|
| cysteine metabolic process | GO:0006534 |  | 2.476E-05 | 0.01138808 |
|---|
| glycolysis | GO:0006096 |  | 2.543E-05 | 9.75E-03 |
|---|
| response to UV | GO:0009411 |  | 2.91E-05 | 9.562E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.463E-05 | 9.957E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 4.614E-05 | 0.01179095 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 4.614E-05 | 0.01061186 |
|---|
| regulation of embryonic development | GO:0045995 |  | 4.614E-05 | 9.647E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.143E-07 | 5.086E-04 |
|---|
| glycolysis | GO:0006096 |  | 3.332E-07 | 3.953E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.59E-07 | 3.631E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 7.601E-07 | 4.509E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.418E-06 | 6.728E-04 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 2.081E-06 | 8.229E-04 |
|---|
| alcohol catabolic process | GO:0046164 |  | 3.241E-06 | 1.099E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.395E-06 | 1.007E-03 |
|---|
| interphase | GO:0051325 |  | 3.718E-06 | 9.803E-04 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 5.918E-06 | 1.404E-03 |
|---|
| cysteine metabolic process | GO:0006534 |  | 2.087E-05 | 4.503E-03 |
|---|



