Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 7083-8-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2026 | 1.170e-02 | 8.953e-03 | 9.876e-02 |
|---|
| IPC-NIBC-129 | 0.2700 | 3.329e-02 | 4.874e-02 | 3.395e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2422 | 1.679e-02 | 2.669e-02 | 3.583e-01 |
|---|
| Loi_GPL570 | 0.1778 | 1.585e-01 | 1.744e-01 | 5.043e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1714 | 1.000e-06 | 9.000e-06 | 1.000e-06 |
|---|
| Parker_GPL1390 | 0.2158 | 7.781e-02 | 9.462e-02 | 4.967e-01 |
|---|
| Parker_GPL887 | 0.0452 | 3.539e-01 | 3.726e-01 | 3.850e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3795 | 8.274e-01 | 7.896e-01 | 9.992e-01 |
|---|
| Schmidt | 0.2119 | 1.853e-02 | 2.132e-02 | 2.220e-01 |
|---|
| Sotiriou | 0.3190 | 1.083e-02 | 8.306e-03 | 1.972e-01 |
|---|
| Van-De-Vijver | 0.2865 | 1.053e-03 | 1.515e-03 | 6.076e-02 |
|---|
| Zhang | 0.2754 | 1.130e-01 | 7.092e-02 | 2.433e-01 |
|---|
| Zhou | 0.4174 | 5.560e-02 | 4.345e-02 | 3.421e-01 |
|---|
Expression data for subnetwork 7083-8-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
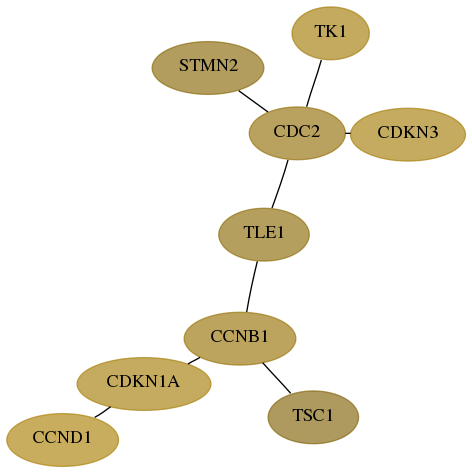
Score for each gene in subnetwork 7083-8-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| cdkn3 |   | 53 | 5 | 29 | 22 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.166 | -0.009 | 0.323 | 0.186 | 0.198 | 0.201 | 0.271 | 0.372 |
|---|
| stmn2 |   | 29 | 9 | 34 | 27 | 0.057 | 0.106 | 0.005 | 0.052 | 0.110 | 0.071 | -0.099 | 0.006 | 0.034 | 0.103 | 0.008 | 0.113 | -0.039 |
|---|
| cdkn1a |   | 44 | 7 | 1 | 4 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | undef | -0.201 | 0.069 |
|---|
| tle1 |   | 47 | 6 | 1 | 3 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | 0.170 | 0.068 | 0.025 |
|---|
| ccnb1 |   | 42 | 8 | 56 | 39 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | undef | 0.167 | 0.398 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| tk1 |   | 7 | 30 | 84 | 67 | 0.109 | 0.149 | 0.190 | -0.017 | 0.131 | 0.105 | -0.100 | 0.257 | 0.204 | 0.142 | 0.293 | 0.164 | 0.222 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
GO Enrichment output for subnetwork 7083-8-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.398E-09 | 1.702E-05 |
|---|
| interphase | GO:0051325 |  | 8.314E-09 | 9.562E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.246E-08 | 3.255E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.563E-08 | 4.924E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 6.734E-07 | 3.098E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.861E-06 | 2.63E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 6.861E-06 | 2.254E-03 |
|---|
| response to UV | GO:0009411 |  | 7.588E-06 | 2.181E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 3.679E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.439E-05 | 3.311E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.785E-05 | 3.733E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.156E-09 | 5.267E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.171E-08 | 1.431E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.824E-08 | 2.299E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.476E-07 | 2.123E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.005E-06 | 1.468E-03 |
|---|
| response to UV | GO:0009411 |  | 3.923E-06 | 1.597E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 4.506E-06 | 1.573E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 7.458E-06 | 2.278E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 7.795E-06 | 2.116E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 8.405E-06 | 2.053E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 8.405E-06 | 1.867E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.328E-09 | 5.602E-06 |
|---|
| interphase | GO:0051325 |  | 2.726E-09 | 3.279E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.434E-08 | 1.15E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.458E-08 | 2.08E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.089E-07 | 1.968E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.37E-06 | 1.351E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 3.37E-06 | 1.158E-03 |
|---|
| response to UV | GO:0009411 |  | 4.121E-06 | 1.239E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 8.768E-06 | 2.344E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 9.163E-06 | 2.205E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.426E-06 | 2.062E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.156E-09 | 5.267E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.171E-08 | 1.431E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.824E-08 | 2.299E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.476E-07 | 2.123E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.005E-06 | 1.468E-03 |
|---|
| response to UV | GO:0009411 |  | 3.923E-06 | 1.597E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 4.506E-06 | 1.573E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 7.458E-06 | 2.278E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 7.795E-06 | 2.116E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 8.405E-06 | 2.053E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 8.405E-06 | 1.867E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.328E-09 | 5.602E-06 |
|---|
| interphase | GO:0051325 |  | 2.726E-09 | 3.279E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.434E-08 | 1.15E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.458E-08 | 2.08E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.089E-07 | 1.968E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.37E-06 | 1.351E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 3.37E-06 | 1.158E-03 |
|---|
| response to UV | GO:0009411 |  | 4.121E-06 | 1.239E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 8.768E-06 | 2.344E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 9.163E-06 | 2.205E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.426E-06 | 2.062E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.608E-10 | 9.313E-07 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.767E-09 | 1.03E-05 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 5.876E-09 | 6.995E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.131E-09 | 7.259E-06 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.73E-08 | 1.236E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 7.119E-08 | 4.237E-05 |
|---|
| response to UV | GO:0009411 |  | 1.913E-06 | 9.76E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.578E-06 | 1.151E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.865E-06 | 1.534E-03 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 4.31E-06 | 1.539E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 5.41E-06 | 1.756E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.315E-10 | 1.209E-06 |
|---|
| interphase | GO:0051325 |  | 3.755E-10 | 6.85E-07 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.016E-09 | 6.099E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 6.139E-09 | 5.598E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.014E-08 | 7.395E-06 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 3.983E-08 | 2.422E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 6.125E-08 | 3.192E-05 |
|---|
| response to UV | GO:0009411 |  | 1.539E-06 | 7.02E-04 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 2.566E-06 | 1.04E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 2.566E-06 | 9.361E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.848E-06 | 1.276E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.328E-09 | 5.602E-06 |
|---|
| interphase | GO:0051325 |  | 2.726E-09 | 3.279E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.434E-08 | 1.15E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.458E-08 | 2.08E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.089E-07 | 1.968E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.37E-06 | 1.351E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 3.37E-06 | 1.158E-03 |
|---|
| response to UV | GO:0009411 |  | 4.121E-06 | 1.239E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 8.768E-06 | 2.344E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 9.163E-06 | 2.205E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.426E-06 | 2.062E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.398E-09 | 1.702E-05 |
|---|
| interphase | GO:0051325 |  | 8.314E-09 | 9.562E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.246E-08 | 3.255E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.563E-08 | 4.924E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 6.734E-07 | 3.098E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.861E-06 | 2.63E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 6.861E-06 | 2.254E-03 |
|---|
| response to UV | GO:0009411 |  | 7.588E-06 | 2.181E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 3.679E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.439E-05 | 3.311E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.785E-05 | 3.733E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.398E-09 | 1.702E-05 |
|---|
| interphase | GO:0051325 |  | 8.314E-09 | 9.562E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.246E-08 | 3.255E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.563E-08 | 4.924E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 6.734E-07 | 3.098E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.861E-06 | 2.63E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 6.861E-06 | 2.254E-03 |
|---|
| response to UV | GO:0009411 |  | 7.588E-06 | 2.181E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 3.679E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.439E-05 | 3.311E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.785E-05 | 3.733E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 6.331E-06 | 0.01166884 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 6.331E-06 | 5.834E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 6.331E-06 | 3.89E-03 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 8.861E-06 | 4.083E-03 |
|---|
| negative regulation of signal transduction | GO:0009968 |  | 1.038E-05 | 3.826E-03 |
|---|
| regulation of stress fiber formation | GO:0051492 |  | 1.181E-05 | 3.628E-03 |
|---|
| negative regulation of cell communication | GO:0010648 |  | 1.275E-05 | 3.358E-03 |
|---|
| regulation of dephosphorylation | GO:0035303 |  | 1.518E-05 | 3.498E-03 |
|---|
| regulation of actin filament bundle formation | GO:0032231 |  | 1.518E-05 | 3.109E-03 |
|---|
| regulation of protein ubiquitination | GO:0031396 |  | 1.897E-05 | 3.497E-03 |
|---|
| negative regulation of translation | GO:0017148 |  | 2.781E-05 | 4.659E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.398E-09 | 1.702E-05 |
|---|
| interphase | GO:0051325 |  | 8.314E-09 | 9.562E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.246E-08 | 3.255E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.563E-08 | 4.924E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 6.734E-07 | 3.098E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.861E-06 | 2.63E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 6.861E-06 | 2.254E-03 |
|---|
| response to UV | GO:0009411 |  | 7.588E-06 | 2.181E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 3.679E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.439E-05 | 3.311E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.785E-05 | 3.733E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.996E-09 | 1.423E-05 |
|---|
| interphase | GO:0051325 |  | 6.73E-09 | 7.985E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.719E-08 | 2.942E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.987E-08 | 4.738E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 5.632E-07 | 2.673E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.06E-06 | 2.397E-03 |
|---|
| response to UV | GO:0009411 |  | 6.779E-06 | 2.298E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 9.085E-06 | 2.695E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.271E-05 | 3.352E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 1.694E-05 | 4.021E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.694E-05 | 3.655E-03 |
|---|



