Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 65108-8-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2198 | 1.678e-02 | 1.347e-02 | 1.307e-01 |
|---|
| IPC-NIBC-129 | 0.2487 | 2.895e-02 | 4.261e-02 | 3.074e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2246 | 7.726e-03 | 1.315e-02 | 2.359e-01 |
|---|
| Loi_GPL570 | 0.2402 | 6.725e-02 | 7.591e-02 | 2.753e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1607 | 3.000e-06 | 3.300e-05 | 9.000e-06 |
|---|
| Parker_GPL1390 | 0.2517 | 8.286e-02 | 1.004e-01 | 5.124e-01 |
|---|
| Parker_GPL887 | 0.1156 | 3.183e-01 | 3.366e-01 | 3.409e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3557 | 4.710e-01 | 3.973e-01 | 9.583e-01 |
|---|
| Schmidt | 0.2075 | 1.630e-02 | 1.859e-02 | 2.032e-01 |
|---|
| Sotiriou | 0.2900 | 1.752e-02 | 1.363e-02 | 2.576e-01 |
|---|
| Van-De-Vijver | 0.2966 | 1.146e-03 | 1.649e-03 | 6.427e-02 |
|---|
| Zhang | 0.2831 | 1.328e-01 | 8.690e-02 | 2.783e-01 |
|---|
| Zhou | 0.4127 | 6.913e-02 | 5.773e-02 | 4.030e-01 |
|---|
Expression data for subnetwork 65108-8-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
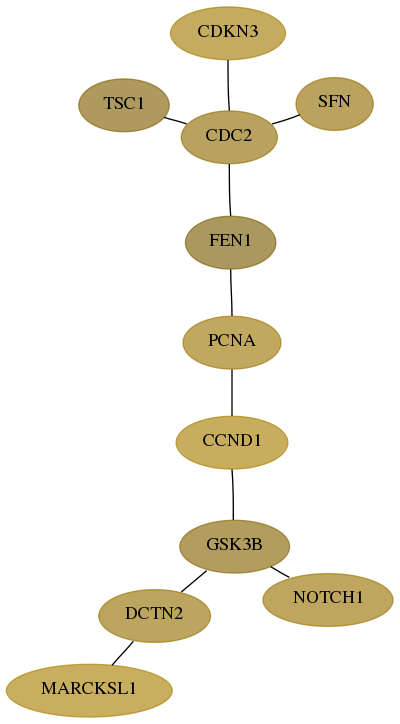
Score for each gene in subnetwork 65108-8-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| cdkn3 |   | 53 | 5 | 29 | 22 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.166 | -0.009 | 0.323 | 0.186 | 0.198 | 0.201 | 0.271 | 0.372 |
|---|
| pcna |   | 15 | 11 | 110 | 85 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.119 | 0.104 | 0.256 | 0.151 | 0.059 | 0.201 | 0.123 | 0.233 |
|---|
| fen1 |   | 9 | 22 | 133 | 112 | 0.045 | 0.194 | 0.169 | 0.196 | 0.136 | 0.252 | 0.033 | 0.332 | 0.221 | 0.144 | 0.242 | 0.056 | 0.114 |
|---|
| notch1 |   | 3 | 62 | 133 | 125 | 0.088 | 0.293 | -0.017 | -0.179 | 0.066 | 0.172 | 0.176 | 0.061 | 0.087 | 0.030 | 0.082 | -0.062 | 0.218 |
|---|
| gsk3b |   | 2 | 78 | 133 | 129 | 0.057 | 0.003 | 0.058 | 0.029 | 0.154 | 0.025 | -0.462 | -0.068 | 0.119 | 0.070 | 0.188 | 0.146 | 0.093 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| dctn2 |   | 1 | 116 | 171 | 176 | 0.085 | 0.090 | 0.096 | 0.052 | 0.158 | 0.110 | -0.170 | 0.156 | -0.003 | 0.136 | 0.064 | 0.032 | 0.199 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| marcksl1 |   | 1 | 116 | 171 | 176 | 0.133 | 0.138 | -0.007 | 0.034 | 0.084 | 0.108 | -0.030 | 0.136 | -0.010 | 0.018 | undef | -0.030 | 0.212 |
|---|
| sfn |   | 61 | 3 | 1 | 2 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.158 | 0.079 | 0.181 |
|---|
GO Enrichment output for subnetwork 65108-8-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.563E-08 | 1.969E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 6.734E-07 | 7.744E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 8.958E-07 | 6.868E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.178E-06 | 3.552E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.861E-06 | 3.156E-03 |
|---|
| anagen | GO:0042640 |  | 6.861E-06 | 2.63E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 6.861E-06 | 2.254E-03 |
|---|
| ectodermal gut morphogenesis | GO:0048567 |  | 6.861E-06 | 1.973E-03 |
|---|
| response to UV | GO:0009411 |  | 7.588E-06 | 1.939E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.029E-05 | 2.366E-03 |
|---|
| ER overload response | GO:0006983 |  | 1.029E-05 | 2.151E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.788E-08 | 4.367E-05 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.814E-07 | 3.438E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 3.952E-07 | 3.218E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.633E-06 | 9.972E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.42E-06 | 1.182E-03 |
|---|
| anagen | GO:0042640 |  | 2.42E-06 | 9.853E-04 |
|---|
| regulation of keratinocyte differentiation | GO:0045616 |  | 2.42E-06 | 8.446E-04 |
|---|
| stem cell division | GO:0017145 |  | 2.42E-06 | 7.39E-04 |
|---|
| response to UV | GO:0009411 |  | 2.814E-06 | 7.638E-04 |
|---|
| hair follicle maturation | GO:0048820 |  | 3.629E-06 | 8.865E-04 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 3.629E-06 | 8.059E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.693E-08 | 6.48E-05 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 3.409E-07 | 4.101E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 4.851E-07 | 3.89E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.234E-06 | 1.344E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.996E-06 | 1.442E-03 |
|---|
| anagen | GO:0042640 |  | 2.996E-06 | 1.201E-03 |
|---|
| regulation of keratinocyte differentiation | GO:0045616 |  | 2.996E-06 | 1.03E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 2.996E-06 | 9.01E-04 |
|---|
| stem cell division | GO:0017145 |  | 2.996E-06 | 8.009E-04 |
|---|
| response to UV | GO:0009411 |  | 3.439E-06 | 8.273E-04 |
|---|
| hair follicle maturation | GO:0048820 |  | 4.492E-06 | 9.826E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.788E-08 | 4.367E-05 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.814E-07 | 3.438E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 3.952E-07 | 3.218E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.633E-06 | 9.972E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.42E-06 | 1.182E-03 |
|---|
| anagen | GO:0042640 |  | 2.42E-06 | 9.853E-04 |
|---|
| regulation of keratinocyte differentiation | GO:0045616 |  | 2.42E-06 | 8.446E-04 |
|---|
| stem cell division | GO:0017145 |  | 2.42E-06 | 7.39E-04 |
|---|
| response to UV | GO:0009411 |  | 2.814E-06 | 7.638E-04 |
|---|
| hair follicle maturation | GO:0048820 |  | 3.629E-06 | 8.865E-04 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 3.629E-06 | 8.059E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.693E-08 | 6.48E-05 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 3.409E-07 | 4.101E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 4.851E-07 | 3.89E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.234E-06 | 1.344E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.996E-06 | 1.442E-03 |
|---|
| anagen | GO:0042640 |  | 2.996E-06 | 1.201E-03 |
|---|
| regulation of keratinocyte differentiation | GO:0045616 |  | 2.996E-06 | 1.03E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 2.996E-06 | 9.01E-04 |
|---|
| stem cell division | GO:0017145 |  | 2.996E-06 | 8.009E-04 |
|---|
| response to UV | GO:0009411 |  | 3.439E-06 | 8.273E-04 |
|---|
| hair follicle maturation | GO:0048820 |  | 4.492E-06 | 9.826E-04 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.259E-08 | 4.494E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.427E-08 | 4.334E-05 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 5.806E-07 | 6.911E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 7.31E-07 | 6.526E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.366E-06 | 2.404E-03 |
|---|
| response to UV | GO:0009411 |  | 4.081E-06 | 2.429E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 4.142E-06 | 2.113E-03 |
|---|
| MutLalpha complex binding | GO:0032405 |  | 4.142E-06 | 1.849E-03 |
|---|
| ribonuclease H activity | GO:0004523 |  | 4.142E-06 | 1.643E-03 |
|---|
| negative regulation of myoblast differentiation | GO:0045662 |  | 4.142E-06 | 1.479E-03 |
|---|
| endodeoxyribonuclease activity. producing 5'-phosphomonoesters | GO:0016888 |  | 4.142E-06 | 1.344E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.125E-08 | 4.104E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.38E-08 | 4.34E-05 |
|---|
| growth cone | GO:0030426 |  | 4.042E-07 | 4.916E-04 |
|---|
| site of polarized growth | GO:0030427 |  | 4.592E-07 | 4.188E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 5.189E-07 | 3.786E-04 |
|---|
| fat cell differentiation | GO:0045444 |  | 5.836E-07 | 3.548E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 7.285E-07 | 3.796E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.448E-06 | 1.116E-03 |
|---|
| response to UV | GO:0009411 |  | 2.813E-06 | 1.14E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 3.763E-06 | 1.373E-03 |
|---|
| ribonuclease H activity | GO:0004523 |  | 3.763E-06 | 1.248E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.693E-08 | 6.48E-05 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 3.409E-07 | 4.101E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 4.851E-07 | 3.89E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.234E-06 | 1.344E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.996E-06 | 1.442E-03 |
|---|
| anagen | GO:0042640 |  | 2.996E-06 | 1.201E-03 |
|---|
| regulation of keratinocyte differentiation | GO:0045616 |  | 2.996E-06 | 1.03E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 2.996E-06 | 9.01E-04 |
|---|
| stem cell division | GO:0017145 |  | 2.996E-06 | 8.009E-04 |
|---|
| response to UV | GO:0009411 |  | 3.439E-06 | 8.273E-04 |
|---|
| hair follicle maturation | GO:0048820 |  | 4.492E-06 | 9.826E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.563E-08 | 1.969E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 6.734E-07 | 7.744E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 8.958E-07 | 6.868E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.178E-06 | 3.552E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.861E-06 | 3.156E-03 |
|---|
| anagen | GO:0042640 |  | 6.861E-06 | 2.63E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 6.861E-06 | 2.254E-03 |
|---|
| ectodermal gut morphogenesis | GO:0048567 |  | 6.861E-06 | 1.973E-03 |
|---|
| response to UV | GO:0009411 |  | 7.588E-06 | 1.939E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.029E-05 | 2.366E-03 |
|---|
| ER overload response | GO:0006983 |  | 1.029E-05 | 2.151E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.563E-08 | 1.969E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 6.734E-07 | 7.744E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 8.958E-07 | 6.868E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.178E-06 | 3.552E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.861E-06 | 3.156E-03 |
|---|
| anagen | GO:0042640 |  | 6.861E-06 | 2.63E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 6.861E-06 | 2.254E-03 |
|---|
| ectodermal gut morphogenesis | GO:0048567 |  | 6.861E-06 | 1.973E-03 |
|---|
| response to UV | GO:0009411 |  | 7.588E-06 | 1.939E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.029E-05 | 2.366E-03 |
|---|
| ER overload response | GO:0006983 |  | 1.029E-05 | 2.151E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| hair follicle maturation | GO:0048820 |  | 1.181E-05 | 0.02176916 |
|---|
| regulation of myoblast differentiation | GO:0045661 |  | 1.181E-05 | 0.01088458 |
|---|
| positive regulation of epidermis development | GO:0045684 |  | 1.181E-05 | 7.256E-03 |
|---|
| regulation of epidermal cell differentiation | GO:0045604 |  | 1.181E-05 | 5.442E-03 |
|---|
| ectodermal gut morphogenesis | GO:0048567 |  | 1.181E-05 | 4.354E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 1.431E-05 | 4.397E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 1.771E-05 | 4.662E-03 |
|---|
| ER overload response | GO:0006983 |  | 1.771E-05 | 4.079E-03 |
|---|
| peptidyl-serine phosphorylation | GO:0018105 |  | 1.771E-05 | 3.626E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.771E-05 | 3.263E-03 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 1.771E-05 | 2.967E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.563E-08 | 1.969E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 6.734E-07 | 7.744E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 8.958E-07 | 6.868E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.178E-06 | 3.552E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.861E-06 | 3.156E-03 |
|---|
| anagen | GO:0042640 |  | 6.861E-06 | 2.63E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 6.861E-06 | 2.254E-03 |
|---|
| ectodermal gut morphogenesis | GO:0048567 |  | 6.861E-06 | 1.973E-03 |
|---|
| response to UV | GO:0009411 |  | 7.588E-06 | 1.939E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.029E-05 | 2.366E-03 |
|---|
| ER overload response | GO:0006983 |  | 1.029E-05 | 2.151E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.042E-07 | 2.474E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.038E-06 | 1.232E-03 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.332E-06 | 1.054E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.723E-06 | 3.988E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.867E-06 | 3.259E-03 |
|---|
| anagen | GO:0042640 |  | 6.867E-06 | 2.716E-03 |
|---|
| negative regulation of myoblast differentiation | GO:0045662 |  | 6.867E-06 | 2.328E-03 |
|---|
| regulation of keratinocyte differentiation | GO:0045616 |  | 6.867E-06 | 2.037E-03 |
|---|
| ectodermal gut morphogenesis | GO:0048567 |  | 6.867E-06 | 1.811E-03 |
|---|
| response to UV | GO:0009411 |  | 8.218E-06 | 1.95E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.029E-05 | 2.221E-03 |
|---|



