Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6449-8-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2106 | 1.696e-02 | 1.362e-02 | 1.317e-01 |
|---|
| IPC-NIBC-129 | 0.2961 | 3.508e-02 | 5.124e-02 | 3.521e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2241 | 6.089e-03 | 1.057e-02 | 2.056e-01 |
|---|
| Loi_GPL570 | 0.2042 | 1.108e-01 | 1.233e-01 | 3.969e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1553 | 0.000e+00 | 2.000e-06 | 0.000e+00 |
|---|
| Parker_GPL1390 | 0.2626 | 8.819e-02 | 1.066e-01 | 5.282e-01 |
|---|
| Parker_GPL887 | 0.1012 | 3.368e-01 | 3.554e-01 | 3.639e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3988 | 5.357e-01 | 4.643e-01 | 9.748e-01 |
|---|
| Schmidt | 0.2030 | 3.391e-02 | 4.055e-02 | 3.298e-01 |
|---|
| Sotiriou | 0.3143 | 1.172e-02 | 9.004e-03 | 2.062e-01 |
|---|
| Van-De-Vijver | 0.2827 | 1.213e-03 | 1.745e-03 | 6.673e-02 |
|---|
| Zhang | 0.2385 | 1.441e-01 | 9.625e-02 | 2.976e-01 |
|---|
| Zhou | 0.4096 | 4.653e-02 | 3.437e-02 | 2.972e-01 |
|---|
Expression data for subnetwork 6449-8-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
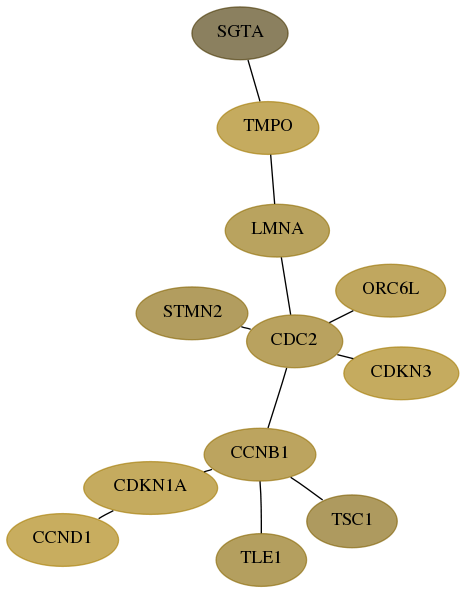
Score for each gene in subnetwork 6449-8-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| cdkn3 |   | 53 | 5 | 29 | 22 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.166 | -0.009 | 0.323 | 0.186 | 0.198 | 0.201 | 0.271 | 0.372 |
|---|
| stmn2 |   | 29 | 9 | 34 | 27 | 0.057 | 0.106 | 0.005 | 0.052 | 0.110 | 0.071 | -0.099 | 0.006 | 0.034 | 0.103 | 0.008 | 0.113 | -0.039 |
|---|
| cdkn1a |   | 44 | 7 | 1 | 4 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | undef | -0.201 | 0.069 |
|---|
| tmpo |   | 2 | 78 | 112 | 109 | 0.116 | 0.177 | 0.104 | 0.256 | 0.163 | 0.152 | 0.068 | 0.284 | 0.099 | 0.144 | 0.148 | 0.130 | 0.309 |
|---|
| tle1 |   | 47 | 6 | 1 | 3 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | 0.170 | 0.068 | 0.025 |
|---|
| lmna |   | 4 | 50 | 132 | 120 | 0.075 | 0.053 | 0.013 | -0.214 | 0.063 | 0.127 | 0.231 | 0.096 | -0.031 | 0.019 | 0.119 | -0.196 | 0.191 |
|---|
| ccnb1 |   | 42 | 8 | 56 | 39 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | undef | 0.167 | 0.398 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| orc6l |   | 15 | 11 | 34 | 28 | 0.094 | 0.270 | 0.084 | 0.114 | 0.106 | 0.240 | 0.256 | 0.234 | 0.192 | 0.105 | 0.258 | 0.175 | 0.160 |
|---|
| sgta |   | 2 | 78 | 112 | 109 | 0.013 | 0.164 | 0.126 | 0.061 | 0.074 | 0.070 | 0.298 | 0.111 | 0.036 | 0.035 | 0.014 | 0.044 | 0.129 |
|---|
GO Enrichment output for subnetwork 6449-8-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 8.037E-08 | 1.849E-04 |
|---|
| interphase | GO:0051325 |  | 9.026E-08 | 1.038E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.715E-07 | 2.082E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.46E-07 | 3.139E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.605E-06 | 1.198E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.652E-05 | 6.332E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.652E-05 | 5.427E-03 |
|---|
| response to UV | GO:0009411 |  | 2.91E-05 | 8.367E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.463E-05 | 8.85E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 3.463E-05 | 7.965E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 4.614E-05 | 9.647E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.939E-08 | 4.737E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.562E-08 | 8.016E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.578E-07 | 1.285E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.232E-06 | 7.526E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.876E-06 | 3.359E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 1.031E-05 | 4.197E-03 |
|---|
| response to UV | GO:0009411 |  | 1.382E-05 | 4.822E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.922E-05 | 5.87E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.922E-05 | 5.218E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 2.47E-05 | 6.034E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.62E-05 | 5.819E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.605E-08 | 6.268E-05 |
|---|
| interphase | GO:0051325 |  | 3.047E-08 | 3.666E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.491E-08 | 7.612E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.281E-07 | 1.372E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.633E-06 | 7.856E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.321E-06 | 3.337E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 8.321E-06 | 2.86E-03 |
|---|
| response to UV | GO:0009411 |  | 1.634E-05 | 4.915E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.326E-05 | 6.218E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.326E-05 | 5.596E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 2.989E-05 | 6.537E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.939E-08 | 4.737E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.562E-08 | 8.016E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.578E-07 | 1.285E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.232E-06 | 7.526E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.876E-06 | 3.359E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 1.031E-05 | 4.197E-03 |
|---|
| response to UV | GO:0009411 |  | 1.382E-05 | 4.822E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.922E-05 | 5.87E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.922E-05 | 5.218E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 2.47E-05 | 6.034E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.62E-05 | 5.819E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.605E-08 | 6.268E-05 |
|---|
| interphase | GO:0051325 |  | 3.047E-08 | 3.666E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.491E-08 | 7.612E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.281E-07 | 1.372E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.633E-06 | 7.856E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.321E-06 | 3.337E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 8.321E-06 | 2.86E-03 |
|---|
| response to UV | GO:0009411 |  | 1.634E-05 | 4.915E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.326E-05 | 6.218E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.326E-05 | 5.596E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 2.989E-05 | 6.537E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.124E-09 | 7.583E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.73E-08 | 3.089E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.7E-08 | 3.214E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.805E-08 | 3.397E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 5.091E-08 | 3.636E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.093E-07 | 1.246E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.061E-06 | 2.582E-03 |
|---|
| response to UV | GO:0009411 |  | 5.598E-06 | 2.499E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.589E-06 | 3.011E-03 |
|---|
| nuclear origin of replication recognition complex | GO:0005664 |  | 7.589E-06 | 2.71E-03 |
|---|
| lamin binding | GO:0005521 |  | 7.589E-06 | 2.464E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.676E-09 | 6.114E-06 |
|---|
| interphase | GO:0051325 |  | 1.898E-09 | 3.462E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.462E-08 | 1.778E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.699E-08 | 1.55E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.43E-08 | 2.503E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 9.481E-08 | 5.764E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.457E-07 | 7.595E-05 |
|---|
| response to UV | GO:0009411 |  | 3.65E-06 | 1.665E-03 |
|---|
| lamin binding | GO:0005521 |  | 4.447E-06 | 1.802E-03 |
|---|
| origin recognition complex | GO:0000808 |  | 4.447E-06 | 1.622E-03 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 4.447E-06 | 1.475E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.605E-08 | 6.268E-05 |
|---|
| interphase | GO:0051325 |  | 3.047E-08 | 3.666E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.491E-08 | 7.612E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.281E-07 | 1.372E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.633E-06 | 7.856E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.321E-06 | 3.337E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 8.321E-06 | 2.86E-03 |
|---|
| response to UV | GO:0009411 |  | 1.634E-05 | 4.915E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.326E-05 | 6.218E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.326E-05 | 5.596E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 2.989E-05 | 6.537E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 8.037E-08 | 1.849E-04 |
|---|
| interphase | GO:0051325 |  | 9.026E-08 | 1.038E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.715E-07 | 2.082E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.46E-07 | 3.139E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.605E-06 | 1.198E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.652E-05 | 6.332E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.652E-05 | 5.427E-03 |
|---|
| response to UV | GO:0009411 |  | 2.91E-05 | 8.367E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.463E-05 | 8.85E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 3.463E-05 | 7.965E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 4.614E-05 | 9.647E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 8.037E-08 | 1.849E-04 |
|---|
| interphase | GO:0051325 |  | 9.026E-08 | 1.038E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.715E-07 | 2.082E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.46E-07 | 3.139E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.605E-06 | 1.198E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.652E-05 | 6.332E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.652E-05 | 5.427E-03 |
|---|
| response to UV | GO:0009411 |  | 2.91E-05 | 8.367E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.463E-05 | 8.85E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 3.463E-05 | 7.965E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 4.614E-05 | 9.647E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.771E-05 | 0.03263475 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 1.771E-05 | 0.01631737 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 1.771E-05 | 0.01087825 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 2.478E-05 | 0.01141552 |
|---|
| regulation of stress fiber formation | GO:0051492 |  | 3.302E-05 | 0.01216947 |
|---|
| regulation of dephosphorylation | GO:0035303 |  | 4.242E-05 | 0.01303113 |
|---|
| regulation of actin filament bundle formation | GO:0032231 |  | 4.242E-05 | 0.01116954 |
|---|
| regulation of protein ubiquitination | GO:0031396 |  | 5.3E-05 | 0.01220958 |
|---|
| negative regulation of signal transduction | GO:0009968 |  | 5.684E-05 | 0.0116399 |
|---|
| negative regulation of cell communication | GO:0010648 |  | 6.972E-05 | 0.01284996 |
|---|
| negative regulation of translation | GO:0017148 |  | 7.764E-05 | 0.0130084 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 8.037E-08 | 1.849E-04 |
|---|
| interphase | GO:0051325 |  | 9.026E-08 | 1.038E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.715E-07 | 2.082E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.46E-07 | 3.139E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.605E-06 | 1.198E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.652E-05 | 6.332E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.652E-05 | 5.427E-03 |
|---|
| response to UV | GO:0009411 |  | 2.91E-05 | 8.367E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.463E-05 | 8.85E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 3.463E-05 | 7.965E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 4.614E-05 | 9.647E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.677E-08 | 1.347E-04 |
|---|
| interphase | GO:0051325 |  | 6.367E-08 | 7.554E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.143E-07 | 1.695E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.59E-07 | 2.723E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.024E-06 | 9.608E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.393E-05 | 5.508E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 2.087E-05 | 7.076E-03 |
|---|
| response to UV | GO:0009411 |  | 2.418E-05 | 7.172E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.92E-05 | 7.699E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.891E-05 | 9.232E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 3.891E-05 | 8.393E-03 |
|---|



