Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6208-7-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1651 | 1.889e-02 | 1.539e-02 | 1.430e-01 |
|---|
| IPC-NIBC-129 | 0.2768 | 3.657e-02 | 5.334e-02 | 3.624e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2189 | 1.478e-02 | 2.377e-02 | 3.357e-01 |
|---|
| Loi_GPL570 | 0.2310 | 7.650e-02 | 8.603e-02 | 3.035e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1585 | 0.000e+00 | 6.000e-06 | 0.000e+00 |
|---|
| Parker_GPL1390 | 0.2217 | 8.144e-02 | 9.880e-02 | 5.080e-01 |
|---|
| Parker_GPL887 | 0.0430 | 4.442e-01 | 4.633e-01 | 4.958e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3752 | 8.388e-01 | 8.030e-01 | 9.994e-01 |
|---|
| Schmidt | 0.2087 | 1.701e-02 | 1.946e-02 | 2.093e-01 |
|---|
| Sotiriou | 0.2738 | 2.290e-02 | 1.797e-02 | 2.973e-01 |
|---|
| Van-De-Vijver | 0.2797 | 7.810e-04 | 1.124e-03 | 4.969e-02 |
|---|
| Zhang | 0.2805 | 1.372e-01 | 9.052e-02 | 2.859e-01 |
|---|
| Zhou | 0.4341 | 5.783e-02 | 4.574e-02 | 3.526e-01 |
|---|
Expression data for subnetwork 6208-7-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
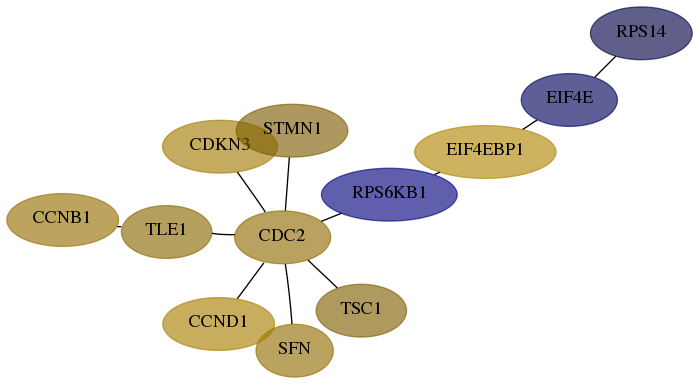
Score for each gene in subnetwork 6208-7-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| cdkn3 |   | 53 | 5 | 29 | 22 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.166 | -0.009 | 0.323 | 0.186 | 0.198 | 0.201 | 0.271 | 0.372 |
|---|
| rps6kb1 |   | 5 | 40 | 216 | 200 | -0.048 | 0.004 | 0.033 | 0.144 | 0.128 | 0.095 | -0.319 | 0.022 | 0.116 | 0.084 | 0.068 | 0.076 | 0.280 |
|---|
| eif4ebp1 |   | 4 | 50 | 260 | 239 | 0.158 | 0.203 | 0.132 | 0.139 | 0.104 | 0.122 | 0.119 | 0.252 | 0.031 | 0.042 | 0.188 | 0.029 | 0.219 |
|---|
| tle1 |   | 47 | 6 | 1 | 3 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | 0.170 | 0.068 | 0.025 |
|---|
| ccnb1 |   | 42 | 8 | 56 | 39 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | undef | 0.167 | 0.398 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| stmn1 |   | 4 | 50 | 173 | 157 | 0.047 | 0.304 | 0.149 | 0.091 | 0.131 | 0.200 | 0.070 | 0.275 | 0.232 | 0.130 | 0.194 | 0.130 | 0.348 |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| rps14 |   | 1 | 116 | 260 | 262 | -0.012 | 0.046 | -0.034 | 0.186 | 0.076 | -0.036 | -0.270 | 0.076 | -0.080 | 0.155 | undef | 0.094 | 0.128 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| eif4e |   | 3 | 62 | 260 | 243 | -0.022 | 0.107 | 0.073 | 0.165 | 0.095 | 0.122 | -0.091 | 0.012 | 0.152 | 0.113 | 0.058 | 0.107 | 0.083 |
|---|
| sfn |   | 61 | 3 | 1 | 2 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.158 | 0.079 | 0.181 |
|---|
GO Enrichment output for subnetwork 6208-7-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of translation | GO:0006417 |  | 1.048E-07 | 2.41E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.706E-07 | 4.262E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 8.282E-07 | 6.349E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.065E-06 | 1.762E-03 |
|---|
| interphase | GO:0051325 |  | 3.36E-06 | 1.546E-03 |
|---|
| translational initiation | GO:0006413 |  | 3.816E-06 | 1.463E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.371E-05 | 4.506E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.371E-05 | 3.943E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.787E-05 | 4.568E-03 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 2.056E-05 | 4.728E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 6.013E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.182E-07 | 2.888E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 4.946E-07 | 6.041E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.057E-06 | 8.609E-04 |
|---|
| translational initiation | GO:0006413 |  | 1.578E-06 | 9.636E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.975E-06 | 2.92E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.496E-06 | 2.645E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 8.959E-06 | 3.127E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.671E-05 | 5.102E-03 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 1.671E-05 | 4.535E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.671E-05 | 4.081E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.12E-05 | 4.708E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.958E-07 | 4.712E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 7.251E-07 | 8.723E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.542E-06 | 1.237E-03 |
|---|
| interphase | GO:0051325 |  | 1.746E-06 | 1.05E-03 |
|---|
| translational initiation | GO:0006413 |  | 2.312E-06 | 1.112E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.727E-06 | 3.098E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 7.727E-06 | 2.656E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.506E-06 | 2.859E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.16E-05 | 5.774E-03 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 2.16E-05 | 5.197E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.16E-05 | 4.724E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.182E-07 | 2.888E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 4.946E-07 | 6.041E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.057E-06 | 8.609E-04 |
|---|
| translational initiation | GO:0006413 |  | 1.578E-06 | 9.636E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.975E-06 | 2.92E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.496E-06 | 2.645E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 8.959E-06 | 3.127E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.671E-05 | 5.102E-03 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 1.671E-05 | 4.535E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.671E-05 | 4.081E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.12E-05 | 4.708E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.958E-07 | 4.712E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 7.251E-07 | 8.723E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.542E-06 | 1.237E-03 |
|---|
| interphase | GO:0051325 |  | 1.746E-06 | 1.05E-03 |
|---|
| translational initiation | GO:0006413 |  | 2.312E-06 | 1.112E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.727E-06 | 3.098E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 7.727E-06 | 2.656E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.506E-06 | 2.859E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.16E-05 | 5.774E-03 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 2.16E-05 | 5.197E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.16E-05 | 4.724E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.805E-08 | 1.359E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 2.511E-07 | 4.483E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.854E-07 | 3.398E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.618E-06 | 4.122E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.061E-06 | 3.615E-03 |
|---|
| eukaryotic initiation factor 4E binding | GO:0008190 |  | 5.061E-06 | 3.012E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.589E-06 | 3.871E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 1.062E-05 | 4.74E-03 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 1.062E-05 | 4.214E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.062E-05 | 3.792E-03 |
|---|
| translation initiation factor binding | GO:0031369 |  | 1.062E-05 | 3.448E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.38E-08 | 8.681E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.568E-07 | 2.86E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 1.633E-07 | 1.986E-04 |
|---|
| interphase | GO:0051325 |  | 1.731E-07 | 1.578E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.448E-06 | 1.786E-03 |
|---|
| response to insulin stimulus | GO:0032868 |  | 3.212E-06 | 1.953E-03 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 3.763E-06 | 1.961E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 3.763E-06 | 1.716E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 5.642E-06 | 2.287E-03 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 5.642E-06 | 2.058E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 5.642E-06 | 1.871E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.958E-07 | 4.712E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 7.251E-07 | 8.723E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.542E-06 | 1.237E-03 |
|---|
| interphase | GO:0051325 |  | 1.746E-06 | 1.05E-03 |
|---|
| translational initiation | GO:0006413 |  | 2.312E-06 | 1.112E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.727E-06 | 3.098E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 7.727E-06 | 2.656E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.506E-06 | 2.859E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.16E-05 | 5.774E-03 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 2.16E-05 | 5.197E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.16E-05 | 4.724E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of translation | GO:0006417 |  | 1.048E-07 | 2.41E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.706E-07 | 4.262E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 8.282E-07 | 6.349E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.065E-06 | 1.762E-03 |
|---|
| interphase | GO:0051325 |  | 3.36E-06 | 1.546E-03 |
|---|
| translational initiation | GO:0006413 |  | 3.816E-06 | 1.463E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.371E-05 | 4.506E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.371E-05 | 3.943E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.787E-05 | 4.568E-03 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 2.056E-05 | 4.728E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 6.013E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of translation | GO:0006417 |  | 1.048E-07 | 2.41E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.706E-07 | 4.262E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 8.282E-07 | 6.349E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.065E-06 | 1.762E-03 |
|---|
| interphase | GO:0051325 |  | 3.36E-06 | 1.546E-03 |
|---|
| translational initiation | GO:0006413 |  | 3.816E-06 | 1.463E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.371E-05 | 4.506E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.371E-05 | 3.943E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.787E-05 | 4.568E-03 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 2.056E-05 | 4.728E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 6.013E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of translation | GO:0017148 |  | 2.26E-07 | 4.165E-04 |
|---|
| regulation of translation | GO:0006417 |  | 6.294E-07 | 5.8E-04 |
|---|
| posttranscriptional regulation of gene expression | GO:0010608 |  | 2.594E-06 | 1.594E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 1.771E-05 | 8.159E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.771E-05 | 6.527E-03 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 1.771E-05 | 5.439E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 1.771E-05 | 4.662E-03 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 2.478E-05 | 5.708E-03 |
|---|
| cellular protein complex disassembly | GO:0043624 |  | 2.478E-05 | 5.074E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 2.478E-05 | 4.566E-03 |
|---|
| regulation of stress fiber formation | GO:0051492 |  | 3.302E-05 | 5.532E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of translation | GO:0006417 |  | 1.048E-07 | 2.41E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.706E-07 | 4.262E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 8.282E-07 | 6.349E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.065E-06 | 1.762E-03 |
|---|
| interphase | GO:0051325 |  | 3.36E-06 | 1.546E-03 |
|---|
| translational initiation | GO:0006413 |  | 3.816E-06 | 1.463E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.371E-05 | 4.506E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.371E-05 | 3.943E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.787E-05 | 4.568E-03 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 2.056E-05 | 4.728E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 6.013E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| ribosomal subunit assembly | GO:0042257 |  | 9.698E-12 | 2.301E-08 |
|---|
| ribosome assembly | GO:0042255 |  | 2.625E-09 | 3.115E-06 |
|---|
| ribosomal small subunit biogenesis | GO:0042274 |  | 5.87E-09 | 4.643E-06 |
|---|
| erythrocyte differentiation | GO:0030218 |  | 7.751E-08 | 4.598E-05 |
|---|
| erythrocyte homeostasis | GO:0034101 |  | 1.721E-07 | 8.167E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.454E-07 | 2.157E-04 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 5.885E-07 | 1.995E-04 |
|---|
| myeloid cell differentiation | GO:0030099 |  | 1.172E-06 | 3.478E-04 |
|---|
| homeostasis of number of cells | GO:0048872 |  | 1.886E-06 | 4.971E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 1.972E-06 | 4.68E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.028E-06 | 8.69E-04 |
|---|



