Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6198-7-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1307 | 1.151e-02 | 8.786e-03 | 9.747e-02 |
|---|
| IPC-NIBC-129 | 0.2632 | 4.923e-02 | 7.090e-02 | 4.407e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2430 | 5.913e-03 | 1.029e-02 | 2.021e-01 |
|---|
| Loi_GPL570 | 0.2490 | 5.942e-02 | 6.730e-02 | 2.503e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1655 | 1.000e-06 | 1.300e-05 | 1.000e-06 |
|---|
| Parker_GPL1390 | 0.2640 | 5.084e-02 | 6.310e-02 | 3.975e-01 |
|---|
| Parker_GPL887 | 0.1319 | 5.433e-01 | 5.616e-01 | 6.138e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3521 | 4.051e-01 | 3.312e-01 | 9.333e-01 |
|---|
| Schmidt | 0.2418 | 1.937e-02 | 2.236e-02 | 2.289e-01 |
|---|
| Sotiriou | 0.2636 | 2.705e-02 | 2.134e-02 | 3.243e-01 |
|---|
| Van-De-Vijver | 0.2581 | 1.459e-03 | 2.099e-03 | 7.540e-02 |
|---|
| Zhang | 0.2726 | 1.236e-01 | 7.941e-02 | 2.623e-01 |
|---|
| Zhou | 0.3992 | 7.144e-02 | 6.025e-02 | 4.127e-01 |
|---|
Expression data for subnetwork 6198-7-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
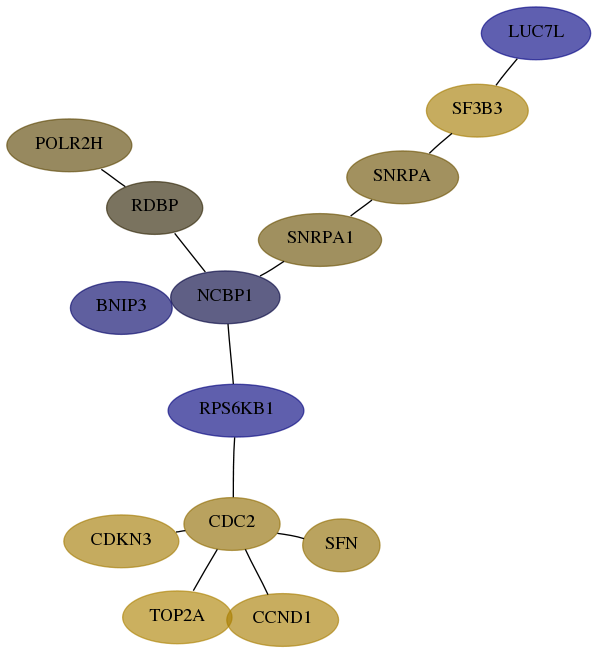
Score for each gene in subnetwork 6198-7-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| cdkn3 |   | 53 | 5 | 29 | 22 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.166 | -0.009 | 0.323 | 0.186 | 0.198 | 0.201 | 0.271 | 0.372 |
|---|
| rdbp |   | 1 | 116 | 216 | 218 | 0.006 | 0.165 | 0.250 | 0.202 | 0.177 | 0.175 | -0.127 | 0.222 | 0.196 | 0.164 | 0.108 | -0.020 | -0.011 |
|---|
| rps6kb1 |   | 5 | 40 | 216 | 200 | -0.048 | 0.004 | 0.033 | 0.144 | 0.128 | 0.095 | -0.319 | 0.022 | 0.116 | 0.084 | 0.068 | 0.076 | 0.280 |
|---|
| luc7l |   | 1 | 116 | 216 | 218 | -0.052 | -0.002 | -0.028 | -0.012 | 0.058 | 0.045 | 0.049 | -0.028 | 0.038 | 0.046 | 0.009 | 0.084 | 0.214 |
|---|
| ncbp1 |   | 1 | 116 | 216 | 218 | -0.010 | 0.114 | 0.082 | -0.018 | 0.057 | 0.125 | 0.001 | 0.245 | -0.015 | 0.012 | 0.054 | -0.043 | 0.340 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| polr2h |   | 2 | 78 | 216 | 215 | 0.021 | 0.282 | 0.144 | 0.302 | 0.127 | 0.204 | 0.069 | 0.129 | 0.257 | 0.153 | undef | 0.165 | 0.236 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| sf3b3 |   | 3 | 62 | 196 | 184 | 0.119 | 0.241 | 0.143 | -0.019 | 0.082 | 0.222 | 0.007 | 0.271 | 0.168 | 0.060 | 0.123 | -0.077 | 0.372 |
|---|
| sfn |   | 61 | 3 | 1 | 2 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.158 | 0.079 | 0.181 |
|---|
| top2a |   | 15 | 11 | 163 | 134 | 0.147 | 0.183 | 0.185 | 0.100 | 0.206 | 0.014 | 0.035 | 0.309 | 0.261 | 0.183 | 0.147 | 0.234 | 0.273 |
|---|
| snrpa1 |   | 1 | 116 | 216 | 218 | 0.029 | 0.139 | 0.128 | 0.087 | 0.093 | 0.310 | -0.081 | 0.201 | 0.180 | 0.072 | undef | 0.090 | 0.169 |
|---|
| snrpa |   | 1 | 116 | 216 | 218 | 0.031 | 0.130 | 0.130 | 0.129 | 0.091 | 0.064 | 0.121 | 0.257 | 0.136 | 0.075 | 0.200 | 0.029 | 0.163 |
|---|
| bnip3 |   | 1 | 116 | 216 | 218 | -0.028 | 0.237 | 0.112 | 0.210 | 0.135 | 0.066 | -0.245 | 0.098 | 0.076 | 0.114 | 0.160 | 0.130 | -0.148 |
|---|
GO Enrichment output for subnetwork 6198-7-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.52E-07 | 1.04E-03 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.654E-06 | 1.903E-03 |
|---|
| nucleus organization | GO:0006997 |  | 9.209E-06 | 7.06E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 1.508E-05 | 8.673E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.508E-05 | 6.938E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.508E-05 | 5.782E-03 |
|---|
| negative regulation of survival gene product expression | GO:0008634 |  | 1.508E-05 | 4.956E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.066E-05 | 5.94E-03 |
|---|
| neuron apoptosis | GO:0051402 |  | 2.261E-05 | 5.777E-03 |
|---|
| DNA topological change | GO:0006265 |  | 2.261E-05 | 5.2E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.163E-05 | 6.613E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of striated muscle development | GO:0045843 |  | 6.484E-08 | 1.584E-04 |
|---|
| negative regulation of muscle development | GO:0048635 |  | 8.911E-08 | 1.088E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.578E-07 | 1.285E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 8.265E-07 | 5.048E-04 |
|---|
| regulation of striated muscle tissue development | GO:0016202 |  | 3.487E-06 | 1.704E-03 |
|---|
| regulation of muscle development | GO:0048634 |  | 3.801E-06 | 1.548E-03 |
|---|
| nucleus organization | GO:0006997 |  | 5.248E-06 | 1.832E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 6.876E-06 | 2.1E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 6.876E-06 | 1.866E-03 |
|---|
| negative regulation of survival gene product expression | GO:0008634 |  | 6.876E-06 | 1.68E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.036E-06 | 1.785E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of striated muscle development | GO:0045843 |  | 7.678E-08 | 1.847E-04 |
|---|
| negative regulation of muscle development | GO:0048635 |  | 1.055E-07 | 1.269E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.958E-07 | 1.571E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 9.783E-07 | 5.884E-04 |
|---|
| regulation of striated muscle tissue development | GO:0016202 |  | 3.774E-06 | 1.816E-03 |
|---|
| regulation of muscle development | GO:0048634 |  | 4.126E-06 | 1.654E-03 |
|---|
| nucleus organization | GO:0006997 |  | 5.308E-06 | 1.824E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 7.727E-06 | 2.324E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 7.727E-06 | 2.066E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 7.727E-06 | 1.859E-03 |
|---|
| negative regulation of survival gene product expression | GO:0008634 |  | 7.727E-06 | 1.69E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of striated muscle development | GO:0045843 |  | 6.484E-08 | 1.584E-04 |
|---|
| negative regulation of muscle development | GO:0048635 |  | 8.911E-08 | 1.088E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.578E-07 | 1.285E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 8.265E-07 | 5.048E-04 |
|---|
| regulation of striated muscle tissue development | GO:0016202 |  | 3.487E-06 | 1.704E-03 |
|---|
| regulation of muscle development | GO:0048634 |  | 3.801E-06 | 1.548E-03 |
|---|
| nucleus organization | GO:0006997 |  | 5.248E-06 | 1.832E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 6.876E-06 | 2.1E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 6.876E-06 | 1.866E-03 |
|---|
| negative regulation of survival gene product expression | GO:0008634 |  | 6.876E-06 | 1.68E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.036E-06 | 1.785E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of striated muscle development | GO:0045843 |  | 7.678E-08 | 1.847E-04 |
|---|
| negative regulation of muscle development | GO:0048635 |  | 1.055E-07 | 1.269E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.958E-07 | 1.571E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 9.783E-07 | 5.884E-04 |
|---|
| regulation of striated muscle tissue development | GO:0016202 |  | 3.774E-06 | 1.816E-03 |
|---|
| regulation of muscle development | GO:0048634 |  | 4.126E-06 | 1.654E-03 |
|---|
| nucleus organization | GO:0006997 |  | 5.308E-06 | 1.824E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 7.727E-06 | 2.324E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 7.727E-06 | 2.066E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 7.727E-06 | 1.859E-03 |
|---|
| negative regulation of survival gene product expression | GO:0008634 |  | 7.727E-06 | 1.69E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.202E-08 | 2.929E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 9.407E-07 | 1.68E-03 |
|---|
| small nuclear ribonucleoprotein complex | GO:0030532 |  | 1.22E-06 | 1.453E-03 |
|---|
| nucleus organization | GO:0006997 |  | 5.203E-06 | 4.645E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 7.176E-06 | 5.125E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 7.176E-06 | 4.271E-03 |
|---|
| negative regulation of survival gene product expression | GO:0008634 |  | 7.176E-06 | 3.661E-03 |
|---|
| alpha-glucosidase activity | GO:0004558 |  | 7.176E-06 | 3.203E-03 |
|---|
| DNA topoisomerase activity | GO:0003916 |  | 7.176E-06 | 2.847E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.969E-06 | 2.846E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.076E-05 | 3.492E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.793E-08 | 1.748E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 4.393E-07 | 8.014E-04 |
|---|
| small nuclear ribonucleoprotein complex | GO:0030532 |  | 6.677E-07 | 8.119E-04 |
|---|
| nucleus organization | GO:0006997 |  | 2.343E-06 | 2.137E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.037E-06 | 2.946E-03 |
|---|
| RS domain binding | GO:0050733 |  | 5.187E-06 | 3.154E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 5.187E-06 | 2.703E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 5.187E-06 | 2.365E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.777E-06 | 3.152E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 7.777E-06 | 2.837E-03 |
|---|
| integral to mitochondrial outer membrane | GO:0031307 |  | 7.777E-06 | 2.579E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of striated muscle development | GO:0045843 |  | 7.678E-08 | 1.847E-04 |
|---|
| negative regulation of muscle development | GO:0048635 |  | 1.055E-07 | 1.269E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.958E-07 | 1.571E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 9.783E-07 | 5.884E-04 |
|---|
| regulation of striated muscle tissue development | GO:0016202 |  | 3.774E-06 | 1.816E-03 |
|---|
| regulation of muscle development | GO:0048634 |  | 4.126E-06 | 1.654E-03 |
|---|
| nucleus organization | GO:0006997 |  | 5.308E-06 | 1.824E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 7.727E-06 | 2.324E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 7.727E-06 | 2.066E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 7.727E-06 | 1.859E-03 |
|---|
| negative regulation of survival gene product expression | GO:0008634 |  | 7.727E-06 | 1.69E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.52E-07 | 1.04E-03 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.654E-06 | 1.903E-03 |
|---|
| nucleus organization | GO:0006997 |  | 9.209E-06 | 7.06E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 1.508E-05 | 8.673E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.508E-05 | 6.938E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.508E-05 | 5.782E-03 |
|---|
| negative regulation of survival gene product expression | GO:0008634 |  | 1.508E-05 | 4.956E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.066E-05 | 5.94E-03 |
|---|
| neuron apoptosis | GO:0051402 |  | 2.261E-05 | 5.777E-03 |
|---|
| DNA topological change | GO:0006265 |  | 2.261E-05 | 5.2E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.163E-05 | 6.613E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.52E-07 | 1.04E-03 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.654E-06 | 1.903E-03 |
|---|
| nucleus organization | GO:0006997 |  | 9.209E-06 | 7.06E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 1.508E-05 | 8.673E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.508E-05 | 6.938E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.508E-05 | 5.782E-03 |
|---|
| negative regulation of survival gene product expression | GO:0008634 |  | 1.508E-05 | 4.956E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.066E-05 | 5.94E-03 |
|---|
| neuron apoptosis | GO:0051402 |  | 2.261E-05 | 5.777E-03 |
|---|
| DNA topological change | GO:0006265 |  | 2.261E-05 | 5.2E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.163E-05 | 6.613E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 2.541E-07 | 4.684E-04 |
|---|
| nucleus organization | GO:0006997 |  | 1.746E-06 | 1.608E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 1.518E-05 | 9.327E-03 |
|---|
| mitochondrion organization | GO:0007005 |  | 1.861E-05 | 8.576E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 2.276E-05 | 8.389E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 3.184E-05 | 9.78E-03 |
|---|
| negative regulation of striated muscle development | GO:0045843 |  | 3.184E-05 | 8.383E-03 |
|---|
| regulation of mRNA processing | GO:0050684 |  | 3.184E-05 | 7.335E-03 |
|---|
| negative regulation of muscle development | GO:0048635 |  | 4.242E-05 | 8.687E-03 |
|---|
| mRNA cleavage | GO:0006379 |  | 4.242E-05 | 7.819E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 4.242E-05 | 7.108E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.52E-07 | 1.04E-03 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.654E-06 | 1.903E-03 |
|---|
| nucleus organization | GO:0006997 |  | 9.209E-06 | 7.06E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 1.508E-05 | 8.673E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.508E-05 | 6.938E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.508E-05 | 5.782E-03 |
|---|
| negative regulation of survival gene product expression | GO:0008634 |  | 1.508E-05 | 4.956E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.066E-05 | 5.94E-03 |
|---|
| neuron apoptosis | GO:0051402 |  | 2.261E-05 | 5.777E-03 |
|---|
| DNA topological change | GO:0006265 |  | 2.261E-05 | 5.2E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.163E-05 | 6.613E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.145E-08 | 2.718E-05 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 3.5E-08 | 4.153E-05 |
|---|
| negative regulation of survival gene product expression | GO:0008634 |  | 3.5E-08 | 2.769E-05 |
|---|
| nucleus organization | GO:0006997 |  | 1.247E-07 | 7.397E-05 |
|---|
| neuron apoptosis | GO:0051402 |  | 1.467E-07 | 6.96E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.454E-07 | 2.157E-04 |
|---|
| regulation of survival gene product expression | GO:0045884 |  | 9.72E-07 | 3.295E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 9.72E-07 | 2.883E-04 |
|---|
| defense response to virus | GO:0051607 |  | 9.72E-07 | 2.563E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.972E-06 | 4.68E-04 |
|---|
| mitochondrion organization | GO:0007005 |  | 2.601E-06 | 5.611E-04 |
|---|



