Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 60-7-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2419 | 1.415e-02 | 1.110e-02 | 1.146e-01 |
|---|
| IPC-NIBC-129 | 0.2606 | 3.072e-02 | 4.511e-02 | 3.208e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2330 | 1.280e-03 | 2.533e-03 | 7.703e-02 |
|---|
| Loi_GPL570 | 0.2906 | 3.280e-02 | 3.773e-02 | 1.549e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1656 | 1.000e-06 | 1.600e-05 | 2.000e-06 |
|---|
| Parker_GPL1390 | 0.3336 | 6.259e-02 | 7.693e-02 | 4.443e-01 |
|---|
| Parker_GPL887 | 0.0807 | 2.773e-01 | 2.948e-01 | 2.904e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3416 | 6.368e-01 | 5.728e-01 | 9.899e-01 |
|---|
| Schmidt | 0.2272 | 8.709e-03 | 9.479e-03 | 1.293e-01 |
|---|
| Sotiriou | 0.3324 | 8.657e-03 | 6.594e-03 | 1.734e-01 |
|---|
| Van-De-Vijver | 0.2923 | 1.222e-03 | 1.759e-03 | 6.709e-02 |
|---|
| Zhang | 0.3210 | 1.234e-01 | 7.924e-02 | 2.619e-01 |
|---|
| Zhou | 0.4091 | 7.863e-02 | 6.821e-02 | 4.418e-01 |
|---|
Expression data for subnetwork 60-7-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
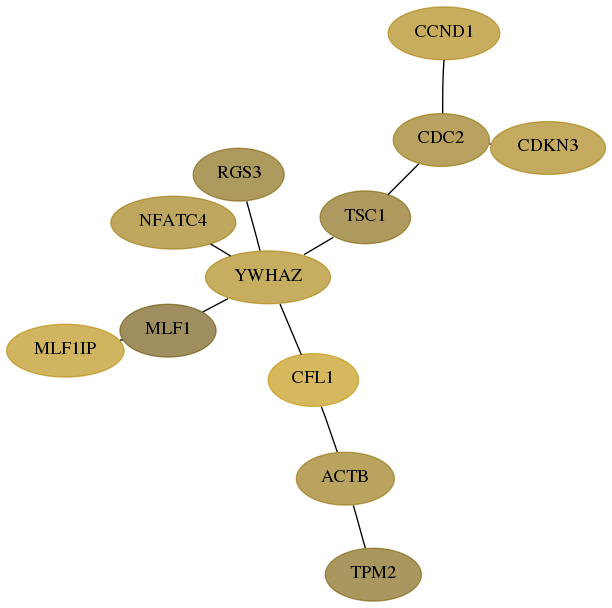
Score for each gene in subnetwork 60-7-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| cdkn3 |   | 53 | 5 | 29 | 22 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.166 | -0.009 | 0.323 | 0.186 | 0.198 | 0.201 | 0.271 | 0.372 |
|---|
| mlf1 |   | 7 | 30 | 56 | 49 | 0.027 | 0.224 | 0.053 | 0.047 | 0.169 | 0.128 | 0.021 | -0.028 | 0.106 | 0.176 | undef | 0.098 | 0.141 |
|---|
| nfatc4 |   | 15 | 11 | 1 | 5 | 0.094 | -0.056 | -0.021 | 0.204 | 0.079 | -0.008 | 0.243 | 0.060 | 0.077 | -0.012 | 0.136 | 0.131 | 0.154 |
|---|
| actb |   | 1 | 116 | 77 | 87 | 0.077 | 0.168 | -0.095 | -0.075 | 0.061 | 0.200 | 0.007 | 0.240 | 0.009 | -0.054 | 0.147 | -0.050 | 0.213 |
|---|
| rgs3 |   | 4 | 50 | 77 | 66 | 0.048 | 0.195 | 0.029 | 0.052 | 0.056 | 0.192 | -0.052 | -0.078 | 0.033 | 0.023 | undef | 0.100 | -0.196 |
|---|
| mlf1ip |   | 5 | 40 | 77 | 64 | 0.182 | 0.147 | 0.186 | 0.144 | 0.242 | 0.266 | 0.005 | 0.345 | 0.182 | 0.197 | undef | 0.240 | 0.372 |
|---|
| ywhaz |   | 17 | 10 | 29 | 25 | 0.124 | 0.089 | 0.087 | 0.200 | 0.141 | 0.110 | 0.228 | 0.199 | 0.056 | 0.123 | undef | 0.103 | 0.213 |
|---|
| cfl1 |   | 8 | 26 | 18 | 16 | 0.216 | 0.134 | 0.083 | -0.084 | 0.142 | 0.251 | -0.466 | 0.229 | 0.174 | 0.188 | 0.208 | -0.025 | 0.215 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| tpm2 |   | 1 | 116 | 77 | 87 | 0.041 | 0.149 | 0.031 | 0.046 | 0.032 | 0.158 | 0.188 | -0.058 | 0.017 | -0.001 | 0.089 | -0.014 | 0.112 |
|---|
GO Enrichment output for subnetwork 60-7-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.269E-08 | 5.219E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.724E-07 | 4.283E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.258E-06 | 9.646E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.552E-06 | 1.468E-03 |
|---|
| sperm motility | GO:0030317 |  | 4.515E-06 | 2.077E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 8.114E-06 | 3.111E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.098E-05 | 3.609E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 1.323E-05 | 3.803E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.802E-05 | 4.604E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.802E-05 | 4.144E-03 |
|---|
| neural plate development | GO:0001840 |  | 1.802E-05 | 3.767E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 8.734E-09 | 2.134E-05 |
|---|
| neural plate morphogenesis | GO:0001839 |  | 8.734E-09 | 1.067E-05 |
|---|
| neural plate development | GO:0001840 |  | 1.746E-08 | 1.422E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 1.912E-07 | 1.168E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.484E-07 | 1.214E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 4.855E-07 | 1.977E-04 |
|---|
| neural crest cell migration | GO:0001755 |  | 7.065E-07 | 2.466E-04 |
|---|
| regulation of actin cytoskeleton organization | GO:0032956 |  | 9.608E-07 | 2.934E-04 |
|---|
| regulation of actin filament-based process | GO:0032970 |  | 1.123E-06 | 3.048E-04 |
|---|
| positive regulation of cytoskeleton organization | GO:0051495 |  | 1.745E-06 | 4.264E-04 |
|---|
| neural crest cell development | GO:0014032 |  | 2.239E-06 | 4.972E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.197E-08 | 2.879E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.402E-07 | 4.093E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 6.647E-07 | 5.331E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.388E-06 | 1.436E-03 |
|---|
| sperm motility | GO:0030317 |  | 2.712E-06 | 1.305E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 5.817E-06 | 2.333E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 8.349E-06 | 2.87E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.162E-05 | 3.494E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 1.162E-05 | 3.106E-03 |
|---|
| neural plate development | GO:0001840 |  | 1.162E-05 | 2.795E-03 |
|---|
| hemopoietic progenitor cell differentiation | GO:0002244 |  | 1.162E-05 | 2.541E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 8.734E-09 | 2.134E-05 |
|---|
| neural plate morphogenesis | GO:0001839 |  | 8.734E-09 | 1.067E-05 |
|---|
| neural plate development | GO:0001840 |  | 1.746E-08 | 1.422E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 1.912E-07 | 1.168E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.484E-07 | 1.214E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 4.855E-07 | 1.977E-04 |
|---|
| neural crest cell migration | GO:0001755 |  | 7.065E-07 | 2.466E-04 |
|---|
| regulation of actin cytoskeleton organization | GO:0032956 |  | 9.608E-07 | 2.934E-04 |
|---|
| regulation of actin filament-based process | GO:0032970 |  | 1.123E-06 | 3.048E-04 |
|---|
| positive regulation of cytoskeleton organization | GO:0051495 |  | 1.745E-06 | 4.264E-04 |
|---|
| neural crest cell development | GO:0014032 |  | 2.239E-06 | 4.972E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.197E-08 | 2.879E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.402E-07 | 4.093E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 6.647E-07 | 5.331E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.388E-06 | 1.436E-03 |
|---|
| sperm motility | GO:0030317 |  | 2.712E-06 | 1.305E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 5.817E-06 | 2.333E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 8.349E-06 | 2.87E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.162E-05 | 3.494E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 1.162E-05 | 3.106E-03 |
|---|
| neural plate development | GO:0001840 |  | 1.162E-05 | 2.795E-03 |
|---|
| hemopoietic progenitor cell differentiation | GO:0002244 |  | 1.162E-05 | 2.541E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to calcium ion | GO:0051592 |  | 1.303E-08 | 4.652E-05 |
|---|
| NuA4 histone acetyltransferase complex | GO:0035267 |  | 1.442E-08 | 2.575E-05 |
|---|
| H4/H2A histone acetyltransferase complex | GO:0043189 |  | 2.306E-08 | 2.745E-05 |
|---|
| nitric-oxide synthase binding | GO:0050998 |  | 4.937E-08 | 4.408E-05 |
|---|
| GTP-dependent protein binding | GO:0030742 |  | 9.043E-08 | 6.459E-05 |
|---|
| histone acetyltransferase complex | GO:0000123 |  | 9.043E-08 | 5.382E-05 |
|---|
| sarcomere organization | GO:0045214 |  | 1.175E-07 | 5.995E-05 |
|---|
| response to metal ion | GO:0010038 |  | 1.663E-07 | 7.422E-05 |
|---|
| response to inorganic substance | GO:0010035 |  | 2.247E-07 | 8.915E-05 |
|---|
| myofibril assembly | GO:0030239 |  | 3.97E-07 | 1.418E-04 |
|---|
| muscle cell development | GO:0055001 |  | 5.445E-07 | 1.768E-04 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.177E-06 | 0.01159071 |
|---|
| neural plate morphogenesis | GO:0001839 |  | 4.447E-06 | 8.111E-03 |
|---|
| hemopoietic progenitor cell differentiation | GO:0002244 |  | 4.447E-06 | 5.407E-03 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 4.447E-06 | 4.055E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 4.447E-06 | 3.244E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.344E-06 | 3.249E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.667E-06 | 3.475E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 6.667E-06 | 3.04E-03 |
|---|
| neural plate development | GO:0001840 |  | 6.667E-06 | 2.702E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.667E-06 | 2.432E-03 |
|---|
| NuA4 histone acetyltransferase complex | GO:0035267 |  | 6.667E-06 | 2.211E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.197E-08 | 2.879E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.402E-07 | 4.093E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 6.647E-07 | 5.331E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.388E-06 | 1.436E-03 |
|---|
| sperm motility | GO:0030317 |  | 2.712E-06 | 1.305E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 5.817E-06 | 2.333E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 8.349E-06 | 2.87E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.162E-05 | 3.494E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 1.162E-05 | 3.106E-03 |
|---|
| neural plate development | GO:0001840 |  | 1.162E-05 | 2.795E-03 |
|---|
| hemopoietic progenitor cell differentiation | GO:0002244 |  | 1.162E-05 | 2.541E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.269E-08 | 5.219E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.724E-07 | 4.283E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.258E-06 | 9.646E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.552E-06 | 1.468E-03 |
|---|
| sperm motility | GO:0030317 |  | 4.515E-06 | 2.077E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 8.114E-06 | 3.111E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.098E-05 | 3.609E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 1.323E-05 | 3.803E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.802E-05 | 4.604E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.802E-05 | 4.144E-03 |
|---|
| neural plate development | GO:0001840 |  | 1.802E-05 | 3.767E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.269E-08 | 5.219E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.724E-07 | 4.283E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.258E-06 | 9.646E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.552E-06 | 1.468E-03 |
|---|
| sperm motility | GO:0030317 |  | 4.515E-06 | 2.077E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 8.114E-06 | 3.111E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.098E-05 | 3.609E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 1.323E-05 | 3.803E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.802E-05 | 4.604E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.802E-05 | 4.144E-03 |
|---|
| neural plate development | GO:0001840 |  | 1.802E-05 | 3.767E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of actin cytoskeleton organization | GO:0032956 |  | 5.159E-06 | 9.509E-03 |
|---|
| regulation of actin filament-based process | GO:0032970 |  | 5.518E-06 | 5.085E-03 |
|---|
| sarcomere organization | GO:0045214 |  | 6.331E-06 | 3.89E-03 |
|---|
| establishment of cell polarity | GO:0030010 |  | 6.331E-06 | 2.917E-03 |
|---|
| regulation of GTPase activity | GO:0043087 |  | 8.017E-06 | 2.955E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 9.493E-06 | 2.916E-03 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 9.493E-06 | 2.499E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 9.493E-06 | 2.187E-03 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 1.329E-05 | 2.721E-03 |
|---|
| myofibril assembly | GO:0030239 |  | 1.329E-05 | 2.449E-03 |
|---|
| regulation of ATPase activity | GO:0043462 |  | 1.329E-05 | 2.226E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.269E-08 | 5.219E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.724E-07 | 4.283E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.258E-06 | 9.646E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.552E-06 | 1.468E-03 |
|---|
| sperm motility | GO:0030317 |  | 4.515E-06 | 2.077E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 8.114E-06 | 3.111E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.098E-05 | 3.609E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 1.323E-05 | 3.803E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.802E-05 | 4.604E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.802E-05 | 4.144E-03 |
|---|
| neural plate development | GO:0001840 |  | 1.802E-05 | 3.767E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.349E-08 | 3.201E-05 |
|---|
| response to calcium ion | GO:0051592 |  | 1.628E-07 | 1.931E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.952E-07 | 2.335E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 9.09E-07 | 5.393E-04 |
|---|
| sarcomere organization | GO:0045214 |  | 1.293E-06 | 6.139E-04 |
|---|
| response to metal ion | GO:0010038 |  | 1.65E-06 | 6.525E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.051E-06 | 6.953E-04 |
|---|
| response to inorganic substance | GO:0010035 |  | 2.131E-06 | 6.322E-04 |
|---|
| sperm motility | GO:0030317 |  | 3.452E-06 | 9.102E-04 |
|---|
| myofibril assembly | GO:0030239 |  | 3.881E-06 | 9.209E-04 |
|---|
| muscle cell development | GO:0055001 |  | 4.841E-06 | 1.044E-03 |
|---|



