Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 55775-6-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2340 | 4.026e-03 | 2.659e-03 | 4.142e-02 |
|---|
| IPC-NIBC-129 | 0.2490 | 3.576e-02 | 5.221e-02 | 3.569e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2938 | 1.197e-02 | 1.961e-02 | 3.004e-01 |
|---|
| Loi_GPL570 | 0.3259 | 1.963e-02 | 2.287e-02 | 9.995e-02 |
|---|
| Loi_GPL96-GPL97 | 0.1540 | 3.000e-06 | 3.200e-05 | 9.000e-06 |
|---|
| Parker_GPL1390 | 0.2315 | 6.985e-02 | 8.539e-02 | 4.703e-01 |
|---|
| Parker_GPL887 | 0.0263 | 2.913e-01 | 3.091e-01 | 3.076e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3355 | 9.224e-01 | 9.036e-01 | 9.999e-01 |
|---|
| Schmidt | 0.2195 | 1.788e-01 | 2.261e-01 | 7.635e-01 |
|---|
| Sotiriou | 0.3523 | 6.204e-03 | 4.681e-03 | 1.427e-01 |
|---|
| Van-De-Vijver | 0.2813 | 7.670e-04 | 1.104e-03 | 4.909e-02 |
|---|
| Zhang | 0.1338 | 1.469e-01 | 9.858e-02 | 3.022e-01 |
|---|
| Zhou | 0.4351 | 8.307e-02 | 7.321e-02 | 4.589e-01 |
|---|
Expression data for subnetwork 55775-6-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
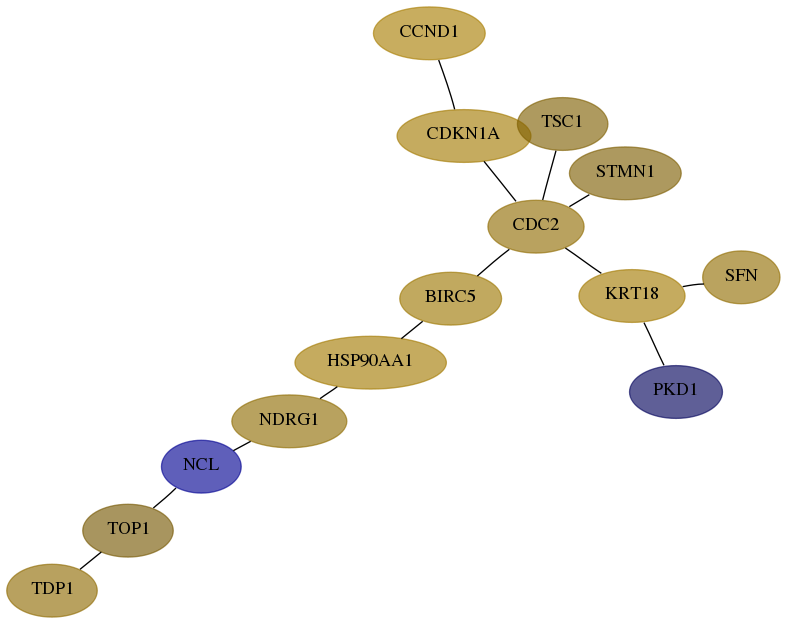
Score for each gene in subnetwork 55775-6-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| ncl |   | 1 | 116 | 184 | 193 | -0.076 | 0.184 | 0.042 | 0.136 | 0.064 | 0.160 | 0.235 | 0.069 | 0.204 | 0.011 | 0.069 | -0.142 | 0.303 |
|---|
| cdkn1a |   | 44 | 7 | 1 | 4 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | undef | -0.201 | 0.069 |
|---|
| top1 |   | 1 | 116 | 184 | 193 | 0.038 | 0.073 | 0.107 | 0.079 | 0.120 | -0.036 | 0.043 | 0.051 | 0.134 | 0.108 | undef | 0.000 | 0.280 |
|---|
| krt18 |   | 6 | 35 | 184 | 166 | 0.112 | -0.078 | 0.147 | 0.150 | 0.161 | -0.086 | -0.204 | 0.083 | -0.099 | 0.118 | -0.024 | -0.114 | 0.222 |
|---|
| tdp1 |   | 1 | 116 | 184 | 193 | 0.072 | 0.099 | 0.101 | 0.012 | 0.068 | 0.089 | 0.152 | 0.281 | 0.066 | 0.080 | 0.038 | 0.053 | 0.238 |
|---|
| hsp90aa1 |   | 5 | 40 | 106 | 90 | 0.116 | 0.030 | 0.164 | 0.076 | 0.119 | 0.155 | -0.199 | 0.169 | 0.069 | 0.102 | undef | -0.011 | 0.399 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| stmn1 |   | 4 | 50 | 173 | 157 | 0.047 | 0.304 | 0.149 | 0.091 | 0.131 | 0.200 | 0.070 | 0.275 | 0.232 | 0.130 | 0.194 | 0.130 | 0.348 |
|---|
| ndrg1 |   | 4 | 50 | 184 | 168 | 0.072 | 0.301 | 0.084 | 0.324 | 0.108 | 0.102 | 0.240 | 0.083 | 0.068 | 0.064 | 0.223 | 0.092 | -0.255 |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| pkd1 |   | 1 | 116 | 184 | 193 | -0.021 | 0.055 | 0.007 | 0.037 | 0.071 | 0.184 | 0.256 | -0.217 | -0.068 | -0.001 | 0.062 | 0.077 | 0.151 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| sfn |   | 61 | 3 | 1 | 2 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.158 | 0.079 | 0.181 |
|---|
| birc5 |   | 5 | 40 | 103 | 89 | 0.101 | 0.216 | 0.190 | 0.134 | 0.223 | 0.149 | 0.166 | 0.202 | 0.236 | 0.241 | undef | 0.205 | 0.400 |
|---|
GO Enrichment output for subnetwork 55775-6-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA topological change | GO:0006265 |  | 3.968E-08 | 9.127E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.46E-07 | 6.279E-04 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 7.173E-07 | 5.499E-04 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 7.173E-07 | 4.124E-04 |
|---|
| negative regulation of peptidase activity | GO:0010466 |  | 1.336E-06 | 6.148E-04 |
|---|
| DNA duplex unwinding | GO:0032508 |  | 1.336E-06 | 5.123E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.605E-06 | 8.559E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.5E-06 | 1.294E-03 |
|---|
| interphase | GO:0051325 |  | 4.933E-06 | 1.261E-03 |
|---|
| negative regulation of hydrolase activity | GO:0051346 |  | 1.058E-05 | 2.433E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.652E-05 | 3.454E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| embryonic cleavage | GO:0040016 |  | 1.785E-12 | 4.361E-09 |
|---|
| DNA topological change | GO:0006265 |  | 2.495E-11 | 3.047E-08 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 8.439E-10 | 6.872E-07 |
|---|
| DNA duplex unwinding | GO:0032508 |  | 1.715E-09 | 1.047E-06 |
|---|
| DNA-dependent DNA replication | GO:0006261 |  | 9.412E-08 | 4.599E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.369E-07 | 5.576E-05 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 1.767E-07 | 6.165E-05 |
|---|
| negative regulation of peptidase activity | GO:0010466 |  | 3.295E-07 | 1.006E-04 |
|---|
| response to drug | GO:0042493 |  | 6.278E-07 | 1.704E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.11E-06 | 2.711E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.224E-06 | 2.718E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| embryonic cleavage | GO:0040016 |  | 2.985E-12 | 7.182E-09 |
|---|
| DNA topological change | GO:0006265 |  | 4.171E-11 | 5.018E-08 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 1.41E-09 | 1.131E-06 |
|---|
| DNA duplex unwinding | GO:0032508 |  | 2.865E-09 | 1.723E-06 |
|---|
| DNA-dependent DNA replication | GO:0006261 |  | 1.23E-07 | 5.919E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.281E-07 | 9.148E-05 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 2.602E-07 | 8.942E-05 |
|---|
| negative regulation of peptidase activity | GO:0010466 |  | 4.851E-07 | 1.459E-04 |
|---|
| response to drug | GO:0042493 |  | 9.942E-07 | 2.658E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.633E-06 | 3.928E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.795E-06 | 3.925E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| embryonic cleavage | GO:0040016 |  | 1.785E-12 | 4.361E-09 |
|---|
| DNA topological change | GO:0006265 |  | 2.495E-11 | 3.047E-08 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 8.439E-10 | 6.872E-07 |
|---|
| DNA duplex unwinding | GO:0032508 |  | 1.715E-09 | 1.047E-06 |
|---|
| DNA-dependent DNA replication | GO:0006261 |  | 9.412E-08 | 4.599E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.369E-07 | 5.576E-05 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 1.767E-07 | 6.165E-05 |
|---|
| negative regulation of peptidase activity | GO:0010466 |  | 3.295E-07 | 1.006E-04 |
|---|
| response to drug | GO:0042493 |  | 6.278E-07 | 1.704E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.11E-06 | 2.711E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.224E-06 | 2.718E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| embryonic cleavage | GO:0040016 |  | 2.985E-12 | 7.182E-09 |
|---|
| DNA topological change | GO:0006265 |  | 4.171E-11 | 5.018E-08 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 1.41E-09 | 1.131E-06 |
|---|
| DNA duplex unwinding | GO:0032508 |  | 2.865E-09 | 1.723E-06 |
|---|
| DNA-dependent DNA replication | GO:0006261 |  | 1.23E-07 | 5.919E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.281E-07 | 9.148E-05 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 2.602E-07 | 8.942E-05 |
|---|
| negative regulation of peptidase activity | GO:0010466 |  | 4.851E-07 | 1.459E-04 |
|---|
| response to drug | GO:0042493 |  | 9.942E-07 | 2.658E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.633E-06 | 3.928E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.795E-06 | 3.925E-04 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.73E-08 | 6.178E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.805E-08 | 6.793E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 5.091E-08 | 6.06E-05 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 1.044E-07 | 9.321E-05 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 1.122E-07 | 8.012E-05 |
|---|
| kinase regulator activity | GO:0019207 |  | 2.213E-07 | 1.317E-04 |
|---|
| protein kinase inhibitor activity | GO:0004860 |  | 1.004E-06 | 5.121E-04 |
|---|
| kinase inhibitor activity | GO:0019210 |  | 1.243E-06 | 5.549E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.759E-06 | 1.491E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.618E-06 | 1.649E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 5.061E-06 | 1.643E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.861E-08 | 6.787E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.793E-08 | 8.742E-05 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 5.475E-08 | 6.657E-05 |
|---|
| midbody | GO:0030496 |  | 7.296E-08 | 6.654E-05 |
|---|
| negative regulation of peptidase activity | GO:0010466 |  | 1.206E-07 | 8.8E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.206E-07 | 7.333E-05 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 1.373E-07 | 7.158E-05 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 1.854E-07 | 8.454E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.854E-07 | 7.515E-05 |
|---|
| kinase regulator activity | GO:0019207 |  | 2.434E-07 | 8.881E-05 |
|---|
| spindle microtubule | GO:0005876 |  | 2.699E-07 | 8.951E-05 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| embryonic cleavage | GO:0040016 |  | 2.985E-12 | 7.182E-09 |
|---|
| DNA topological change | GO:0006265 |  | 4.171E-11 | 5.018E-08 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 1.41E-09 | 1.131E-06 |
|---|
| DNA duplex unwinding | GO:0032508 |  | 2.865E-09 | 1.723E-06 |
|---|
| DNA-dependent DNA replication | GO:0006261 |  | 1.23E-07 | 5.919E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.281E-07 | 9.148E-05 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 2.602E-07 | 8.942E-05 |
|---|
| negative regulation of peptidase activity | GO:0010466 |  | 4.851E-07 | 1.459E-04 |
|---|
| response to drug | GO:0042493 |  | 9.942E-07 | 2.658E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.633E-06 | 3.928E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.795E-06 | 3.925E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA topological change | GO:0006265 |  | 3.968E-08 | 9.127E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.46E-07 | 6.279E-04 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 7.173E-07 | 5.499E-04 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 7.173E-07 | 4.124E-04 |
|---|
| negative regulation of peptidase activity | GO:0010466 |  | 1.336E-06 | 6.148E-04 |
|---|
| DNA duplex unwinding | GO:0032508 |  | 1.336E-06 | 5.123E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.605E-06 | 8.559E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.5E-06 | 1.294E-03 |
|---|
| interphase | GO:0051325 |  | 4.933E-06 | 1.261E-03 |
|---|
| negative regulation of hydrolase activity | GO:0051346 |  | 1.058E-05 | 2.433E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.652E-05 | 3.454E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA topological change | GO:0006265 |  | 3.968E-08 | 9.127E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.46E-07 | 6.279E-04 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 7.173E-07 | 5.499E-04 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 7.173E-07 | 4.124E-04 |
|---|
| negative regulation of peptidase activity | GO:0010466 |  | 1.336E-06 | 6.148E-04 |
|---|
| DNA duplex unwinding | GO:0032508 |  | 1.336E-06 | 5.123E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.605E-06 | 8.559E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.5E-06 | 1.294E-03 |
|---|
| interphase | GO:0051325 |  | 4.933E-06 | 1.261E-03 |
|---|
| negative regulation of hydrolase activity | GO:0051346 |  | 1.058E-05 | 2.433E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.652E-05 | 3.454E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell growth | GO:0030307 |  | 1.771E-05 | 0.03263475 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.771E-05 | 0.01631737 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 1.771E-05 | 0.01087825 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 1.771E-05 | 8.159E-03 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 2.478E-05 | 9.132E-03 |
|---|
| cellular protein complex disassembly | GO:0043624 |  | 2.478E-05 | 7.61E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 2.478E-05 | 6.523E-03 |
|---|
| regulation of stress fiber formation | GO:0051492 |  | 3.302E-05 | 7.606E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 3.302E-05 | 6.761E-03 |
|---|
| positive regulation of cell size | GO:0045793 |  | 4.242E-05 | 7.819E-03 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 4.242E-05 | 7.108E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA topological change | GO:0006265 |  | 3.968E-08 | 9.127E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.46E-07 | 6.279E-04 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 7.173E-07 | 5.499E-04 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 7.173E-07 | 4.124E-04 |
|---|
| negative regulation of peptidase activity | GO:0010466 |  | 1.336E-06 | 6.148E-04 |
|---|
| DNA duplex unwinding | GO:0032508 |  | 1.336E-06 | 5.123E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.605E-06 | 8.559E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.5E-06 | 1.294E-03 |
|---|
| interphase | GO:0051325 |  | 4.933E-06 | 1.261E-03 |
|---|
| negative regulation of hydrolase activity | GO:0051346 |  | 1.058E-05 | 2.433E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.652E-05 | 3.454E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| detection of mechanical stimulus | GO:0050982 |  | 2.227E-09 | 5.285E-06 |
|---|
| cartilage condensation | GO:0001502 |  | 3.115E-09 | 3.696E-06 |
|---|
| protein refolding | GO:0042026 |  | 4.984E-08 | 3.942E-05 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 8.714E-08 | 5.17E-05 |
|---|
| DNA topological change | GO:0006265 |  | 8.714E-08 | 4.136E-05 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.393E-07 | 5.509E-05 |
|---|
| response to mechanical stimulus | GO:0009612 |  | 1.415E-07 | 4.799E-05 |
|---|
| cell cycle arrest | GO:0007050 |  | 1.81E-07 | 5.37E-05 |
|---|
| JAK-STAT cascade | GO:0007259 |  | 4.104E-07 | 1.082E-04 |
|---|
| detection of abiotic stimulus | GO:0009582 |  | 5.859E-07 | 1.39E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.775E-07 | 1.893E-04 |
|---|



