Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 55367-6-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1898 | 3.476e-02 | 3.057e-02 | 2.241e-01 |
|---|
| IPC-NIBC-129 | 0.2574 | 4.592e-02 | 6.634e-02 | 4.216e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.1889 | 1.830e-04 | 4.230e-04 | 1.940e-02 |
|---|
| Loi_GPL570 | 0.3480 | 1.419e-02 | 1.666e-02 | 7.508e-02 |
|---|
| Loi_GPL96-GPL97 | 0.1294 | 2.000e-06 | 1.900e-05 | 3.000e-06 |
|---|
| Parker_GPL1390 | 0.4213 | 1.111e-02 | 1.475e-02 | 1.578e-01 |
|---|
| Parker_GPL887 | 0.2854 | 3.827e-01 | 4.017e-01 | 4.206e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3359 | 8.222e-02 | 4.931e-02 | 4.793e-01 |
|---|
| Schmidt | 0.3463 | 9.577e-02 | 1.199e-01 | 5.870e-01 |
|---|
| Sotiriou | 0.2946 | 1.625e-02 | 1.261e-02 | 2.472e-01 |
|---|
| Van-De-Vijver | 0.2631 | 2.012e-03 | 2.892e-03 | 9.286e-02 |
|---|
| Zhang | 0.1739 | 2.115e-01 | 1.557e-01 | 4.044e-01 |
|---|
| Zhou | 0.3812 | 8.274e-02 | 7.284e-02 | 4.577e-01 |
|---|
Expression data for subnetwork 55367-6-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
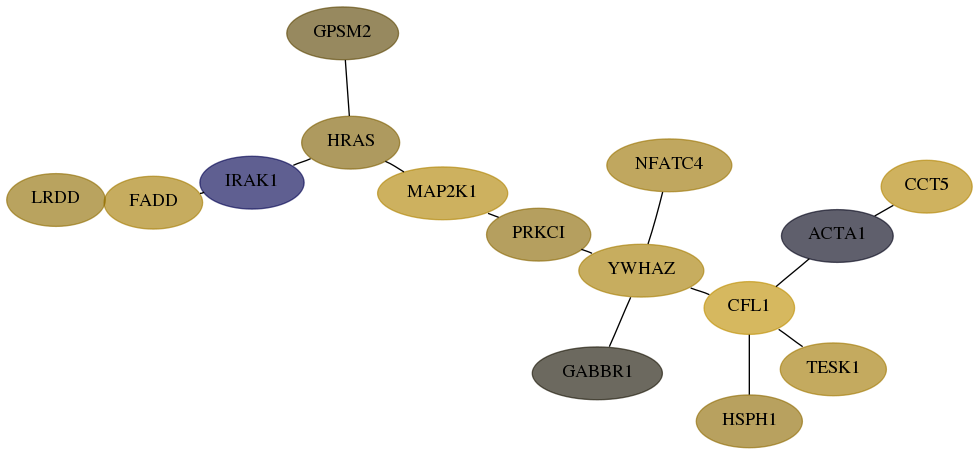
Score for each gene in subnetwork 55367-6-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| nfatc4 |   | 15 | 11 | 1 | 5 | 0.094 | -0.056 | -0.021 | 0.204 | 0.079 | -0.008 | 0.243 | 0.060 | 0.077 | -0.012 | 0.136 | 0.131 | 0.154 |
|---|
| irak1 |   | 5 | 40 | 42 | 36 | -0.016 | 0.242 | 0.007 | 0.133 | 0.108 | 0.303 | 0.257 | 0.133 | 0.177 | 0.084 | 0.199 | -0.026 | 0.081 |
|---|
| ywhaz |   | 17 | 10 | 29 | 25 | 0.124 | 0.089 | 0.087 | 0.200 | 0.141 | 0.110 | 0.228 | 0.199 | 0.056 | 0.123 | undef | 0.103 | 0.213 |
|---|
| cct5 |   | 2 | 78 | 64 | 63 | 0.165 | 0.258 | 0.182 | 0.155 | 0.209 | 0.212 | 0.257 | 0.353 | 0.143 | 0.204 | undef | 0.039 | 0.289 |
|---|
| cfl1 |   | 8 | 26 | 18 | 16 | 0.216 | 0.134 | 0.083 | -0.084 | 0.142 | 0.251 | -0.466 | 0.229 | 0.174 | 0.188 | 0.208 | -0.025 | 0.215 |
|---|
| prkci |   | 14 | 15 | 31 | 26 | 0.065 | 0.104 | 0.012 | 0.170 | 0.112 | 0.088 | -0.103 | -0.170 | 0.196 | 0.128 | 0.049 | 0.124 | 0.149 |
|---|
| fadd |   | 4 | 50 | 42 | 40 | 0.121 | -0.009 | 0.155 | 0.307 | 0.166 | 0.119 | 0.084 | 0.300 | 0.070 | 0.116 | 0.051 | 0.032 | 0.243 |
|---|
| gpsm2 |   | 4 | 50 | 42 | 40 | 0.021 | 0.286 | 0.155 | 0.144 | 0.108 | 0.242 | 0.307 | 0.134 | 0.234 | 0.077 | 0.112 | 0.098 | 0.207 |
|---|
| hras |   | 14 | 15 | 42 | 34 | 0.049 | -0.030 | 0.094 | 0.210 | 0.160 | 0.195 | 0.022 | 0.245 | 0.086 | 0.208 | 0.180 | 0.003 | 0.197 |
|---|
| hsph1 |   | 8 | 26 | 18 | 16 | 0.071 | 0.260 | 0.138 | 0.050 | 0.148 | 0.197 | -0.116 | 0.161 | 0.143 | 0.132 | -0.006 | 0.094 | 0.357 |
|---|
| gabbr1 |   | 7 | 30 | 56 | 49 | 0.003 | -0.016 | -0.022 | -0.051 | 0.082 | 0.119 | 0.584 | -0.082 | 0.049 | 0.057 | 0.125 | 0.096 | 0.116 |
|---|
| acta1 |   | 9 | 22 | 1 | 7 | -0.003 | 0.098 | -0.008 | -0.030 | 0.119 | 0.174 | 0.376 | 0.024 | 0.182 | 0.214 | 0.100 | 0.048 | 0.201 |
|---|
| lrdd |   | 3 | 62 | 42 | 45 | 0.075 | 0.117 | -0.015 | 0.179 | 0.068 | 0.006 | 0.302 | 0.104 | 0.152 | 0.007 | undef | 0.108 | 0.085 |
|---|
| tesk1 |   | 7 | 30 | 18 | 19 | 0.110 | 0.169 | 0.161 | 0.179 | 0.096 | 0.138 | -0.154 | 0.245 | 0.155 | 0.092 | 0.067 | 0.029 | 0.200 |
|---|
| map2k1 |   | 10 | 20 | 54 | 44 | 0.154 | 0.096 | 0.093 | 0.013 | 0.095 | 0.045 | -0.223 | 0.207 | 0.082 | 0.096 | undef | undef | 0.230 |
|---|
GO Enrichment output for subnetwork 55367-6-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.915E-08 | 6.703E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 2.439E-07 | 2.804E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.781E-07 | 3.665E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.615E-06 | 9.283E-04 |
|---|
| actin filament organization | GO:0007015 |  | 1.655E-06 | 7.611E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 3.274E-06 | 1.255E-03 |
|---|
| sperm motility | GO:0030317 |  | 5.789E-06 | 1.902E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.04E-05 | 2.99E-03 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 1.368E-05 | 3.496E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.408E-05 | 3.237E-03 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 1.695E-05 | 3.544E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| establishment of cell polarity | GO:0030010 |  | 2.664E-10 | 6.507E-07 |
|---|
| actin filament organization | GO:0007015 |  | 5.714E-09 | 6.979E-06 |
|---|
| polyol catabolic process | GO:0046174 |  | 6.62E-09 | 5.391E-06 |
|---|
| neural plate morphogenesis | GO:0001839 |  | 6.62E-09 | 4.043E-06 |
|---|
| neural plate development | GO:0001840 |  | 1.323E-08 | 6.465E-06 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 5.35E-08 | 2.179E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.884E-07 | 6.575E-05 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.682E-07 | 1.124E-04 |
|---|
| neural crest cell migration | GO:0001755 |  | 5.359E-07 | 1.455E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 8.718E-07 | 2.13E-04 |
|---|
| positive regulation of cytoskeleton organization | GO:0051495 |  | 1.324E-06 | 2.941E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 8.016E-09 | 1.929E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 9.588E-08 | 1.153E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.281E-07 | 1.829E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 4.457E-07 | 2.681E-04 |
|---|
| actin filament organization | GO:0007015 |  | 4.793E-07 | 2.306E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 9.05E-07 | 3.629E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.603E-06 | 5.508E-04 |
|---|
| sperm motility | GO:0030317 |  | 1.82E-06 | 5.474E-04 |
|---|
| osteoblast differentiation | GO:0001649 |  | 1.82E-06 | 4.865E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 3.907E-06 | 9.4E-04 |
|---|
| negative regulation of adenylate cyclase activity | GO:0007194 |  | 5.61E-06 | 1.227E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| establishment of cell polarity | GO:0030010 |  | 2.664E-10 | 6.507E-07 |
|---|
| actin filament organization | GO:0007015 |  | 5.714E-09 | 6.979E-06 |
|---|
| polyol catabolic process | GO:0046174 |  | 6.62E-09 | 5.391E-06 |
|---|
| neural plate morphogenesis | GO:0001839 |  | 6.62E-09 | 4.043E-06 |
|---|
| neural plate development | GO:0001840 |  | 1.323E-08 | 6.465E-06 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 5.35E-08 | 2.179E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.884E-07 | 6.575E-05 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.682E-07 | 1.124E-04 |
|---|
| neural crest cell migration | GO:0001755 |  | 5.359E-07 | 1.455E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 8.718E-07 | 2.13E-04 |
|---|
| positive regulation of cytoskeleton organization | GO:0051495 |  | 1.324E-06 | 2.941E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 8.016E-09 | 1.929E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 9.588E-08 | 1.153E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.281E-07 | 1.829E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 4.457E-07 | 2.681E-04 |
|---|
| actin filament organization | GO:0007015 |  | 4.793E-07 | 2.306E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 9.05E-07 | 3.629E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.603E-06 | 5.508E-04 |
|---|
| sperm motility | GO:0030317 |  | 1.82E-06 | 5.474E-04 |
|---|
| osteoblast differentiation | GO:0001649 |  | 1.82E-06 | 4.865E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 3.907E-06 | 9.4E-04 |
|---|
| negative regulation of adenylate cyclase activity | GO:0007194 |  | 5.61E-06 | 1.227E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| death receptor binding | GO:0005123 |  | 4.766E-08 | 1.702E-04 |
|---|
| establishment of cell polarity | GO:0030010 |  | 1.02E-07 | 1.821E-04 |
|---|
| actin filament organization | GO:0007015 |  | 2.476E-07 | 2.947E-04 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 2.473E-06 | 2.208E-03 |
|---|
| tumor necrosis factor receptor superfamily binding | GO:0032813 |  | 3.07E-06 | 2.193E-03 |
|---|
| striated muscle cell differentiation | GO:0051146 |  | 3.772E-06 | 2.245E-03 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 4.573E-06 | 2.333E-03 |
|---|
| skeletal myofibril assembly | GO:0014866 |  | 9.657E-06 | 4.311E-03 |
|---|
| neural plate development | GO:0001840 |  | 9.657E-06 | 3.832E-03 |
|---|
| cell cortex part | GO:0044448 |  | 1.347E-05 | 4.812E-03 |
|---|
| lipopolysaccharide-mediated signaling pathway | GO:0031663 |  | 1.448E-05 | 4.7E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| death receptor binding | GO:0005123 |  | 2.325E-08 | 8.482E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 3.486E-08 | 6.359E-05 |
|---|
| actin filament organization | GO:0007015 |  | 1.183E-07 | 1.439E-04 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 7.471E-07 | 6.814E-04 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 1.502E-06 | 1.096E-03 |
|---|
| striated muscle cell differentiation | GO:0051146 |  | 1.502E-06 | 9.132E-04 |
|---|
| tumor necrosis factor receptor superfamily binding | GO:0032813 |  | 1.502E-06 | 7.827E-04 |
|---|
| muscle thin filament assembly | GO:0030240 |  | 5.984E-06 | 2.729E-03 |
|---|
| neural plate morphogenesis | GO:0001839 |  | 5.984E-06 | 2.426E-03 |
|---|
| NF-kappaB-inducing kinase activity | GO:0004704 |  | 5.984E-06 | 2.183E-03 |
|---|
| cell cortex part | GO:0044448 |  | 7.045E-06 | 2.336E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 8.016E-09 | 1.929E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 9.588E-08 | 1.153E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.281E-07 | 1.829E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 4.457E-07 | 2.681E-04 |
|---|
| actin filament organization | GO:0007015 |  | 4.793E-07 | 2.306E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 9.05E-07 | 3.629E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.603E-06 | 5.508E-04 |
|---|
| sperm motility | GO:0030317 |  | 1.82E-06 | 5.474E-04 |
|---|
| osteoblast differentiation | GO:0001649 |  | 1.82E-06 | 4.865E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 3.907E-06 | 9.4E-04 |
|---|
| negative regulation of adenylate cyclase activity | GO:0007194 |  | 5.61E-06 | 1.227E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.915E-08 | 6.703E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 2.439E-07 | 2.804E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.781E-07 | 3.665E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.615E-06 | 9.283E-04 |
|---|
| actin filament organization | GO:0007015 |  | 1.655E-06 | 7.611E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 3.274E-06 | 1.255E-03 |
|---|
| sperm motility | GO:0030317 |  | 5.789E-06 | 1.902E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.04E-05 | 2.99E-03 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 1.368E-05 | 3.496E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.408E-05 | 3.237E-03 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 1.695E-05 | 3.544E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.915E-08 | 6.703E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 2.439E-07 | 2.804E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.781E-07 | 3.665E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.615E-06 | 9.283E-04 |
|---|
| actin filament organization | GO:0007015 |  | 1.655E-06 | 7.611E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 3.274E-06 | 1.255E-03 |
|---|
| sperm motility | GO:0030317 |  | 5.789E-06 | 1.902E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.04E-05 | 2.99E-03 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 1.368E-05 | 3.496E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.408E-05 | 3.237E-03 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 1.695E-05 | 3.544E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| establishment of cell polarity | GO:0030010 |  | 3.036E-08 | 5.595E-05 |
|---|
| actin filament organization | GO:0007015 |  | 6.641E-07 | 6.12E-04 |
|---|
| striated muscle cell differentiation | GO:0051146 |  | 1.369E-06 | 8.412E-04 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 3.416E-06 | 1.574E-03 |
|---|
| muscle cell differentiation | GO:0042692 |  | 1.771E-05 | 6.529E-03 |
|---|
| regulation of long-term neuronal synaptic plasticity | GO:0048169 |  | 2.318E-05 | 7.121E-03 |
|---|
| establishment or maintenance of epithelial cell apical/basal polarity | GO:0045197 |  | 2.318E-05 | 6.103E-03 |
|---|
| activation of NF-kappaB-inducing kinase activity | GO:0007250 |  | 2.318E-05 | 5.34E-03 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 2.318E-05 | 4.747E-03 |
|---|
| establishment or maintenance of apical/basal cell polarity | GO:0035088 |  | 3.474E-05 | 6.403E-03 |
|---|
| regulation of synaptic transmission. GABAergic | GO:0032228 |  | 3.474E-05 | 5.821E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.915E-08 | 6.703E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 2.439E-07 | 2.804E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.781E-07 | 3.665E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.615E-06 | 9.283E-04 |
|---|
| actin filament organization | GO:0007015 |  | 1.655E-06 | 7.611E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 3.274E-06 | 1.255E-03 |
|---|
| sperm motility | GO:0030317 |  | 5.789E-06 | 1.902E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.04E-05 | 2.99E-03 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 1.368E-05 | 3.496E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.408E-05 | 3.237E-03 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 1.695E-05 | 3.544E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.752E-08 | 4.157E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 2.093E-07 | 2.484E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.831E-07 | 3.031E-04 |
|---|
| actin filament organization | GO:0007015 |  | 1.172E-06 | 6.955E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.179E-06 | 5.597E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.66E-06 | 1.052E-03 |
|---|
| sperm motility | GO:0030317 |  | 4.476E-06 | 1.517E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 7.706E-06 | 2.286E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.023E-05 | 2.698E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 1.325E-05 | 3.145E-03 |
|---|
| skeletal myofibril assembly | GO:0014866 |  | 1.514E-05 | 3.265E-03 |
|---|



