Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 54995-6-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1911 | 1.410e-02 | 1.106e-02 | 1.143e-01 |
|---|
| IPC-NIBC-129 | 0.2880 | 4.682e-02 | 6.758e-02 | 4.269e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2331 | 2.359e-02 | 3.639e-02 | 4.235e-01 |
|---|
| Loi_GPL570 | 0.2051 | 1.094e-01 | 1.218e-01 | 3.933e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1626 | 0.000e+00 | 3.000e-06 | 0.000e+00 |
|---|
| Parker_GPL1390 | 0.2000 | 4.374e-02 | 5.468e-02 | 3.658e-01 |
|---|
| Parker_GPL887 | 0.0399 | 3.797e-01 | 3.986e-01 | 4.168e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3664 | 8.552e-01 | 8.224e-01 | 9.996e-01 |
|---|
| Schmidt | 0.2523 | 6.392e-03 | 6.785e-03 | 1.023e-01 |
|---|
| Sotiriou | 0.3234 | 1.007e-02 | 7.702e-03 | 1.891e-01 |
|---|
| Van-De-Vijver | 0.2617 | 1.320e-03 | 1.900e-03 | 7.060e-02 |
|---|
| Zhang | 0.3395 | 1.290e-01 | 8.377e-02 | 2.717e-01 |
|---|
| Zhou | 0.4048 | 6.271e-02 | 5.085e-02 | 3.750e-01 |
|---|
Expression data for subnetwork 54995-6-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
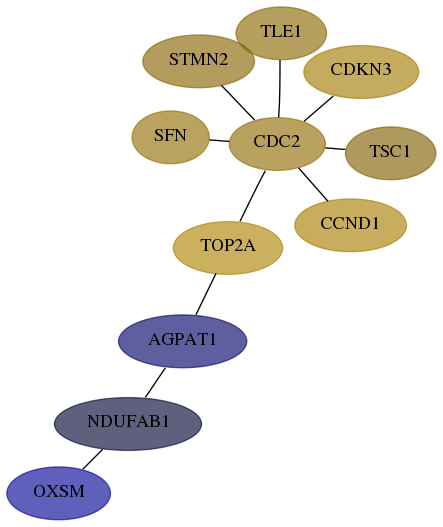
Score for each gene in subnetwork 54995-6-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| cdkn3 |   | 53 | 5 | 29 | 22 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.166 | -0.009 | 0.323 | 0.186 | 0.198 | 0.201 | 0.271 | 0.372 |
|---|
| oxsm |   | 1 | 116 | 193 | 201 | -0.082 | 0.116 | 0.115 | 0.091 | 0.102 | -0.082 | -0.426 | 0.087 | -0.027 | 0.091 | -0.071 | 0.132 | 0.221 |
|---|
| stmn2 |   | 29 | 9 | 34 | 27 | 0.057 | 0.106 | 0.005 | 0.052 | 0.110 | 0.071 | -0.099 | 0.006 | 0.034 | 0.103 | 0.008 | 0.113 | -0.039 |
|---|
| tle1 |   | 47 | 6 | 1 | 3 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | 0.170 | 0.068 | 0.025 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| ndufab1 |   | 2 | 78 | 190 | 182 | -0.007 | 0.205 | 0.083 | 0.106 | 0.157 | 0.038 | -0.073 | 0.175 | 0.193 | 0.142 | 0.079 | 0.031 | 0.138 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| agpat1 |   | 2 | 78 | 190 | 182 | -0.027 | 0.104 | 0.123 | 0.096 | 0.093 | 0.167 | 0.097 | 0.028 | 0.097 | 0.101 | 0.114 | 0.018 | -0.007 |
|---|
| sfn |   | 61 | 3 | 1 | 2 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.158 | 0.079 | 0.181 |
|---|
| top2a |   | 15 | 11 | 163 | 134 | 0.147 | 0.183 | 0.185 | 0.100 | 0.206 | 0.014 | 0.035 | 0.309 | 0.261 | 0.183 | 0.147 | 0.234 | 0.273 |
|---|
GO Enrichment output for subnetwork 54995-6-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.411E-07 | 5.544E-04 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 2.785E-06 | 3.203E-03 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 6.376E-06 | 4.888E-03 |
|---|
| JAK-STAT cascade | GO:0007259 |  | 1.048E-05 | 6.028E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.117E-05 | 5.138E-03 |
|---|
| short-chain fatty acid metabolic process | GO:0046459 |  | 1.117E-05 | 4.282E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.117E-05 | 3.67E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.117E-05 | 3.211E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.117E-05 | 2.854E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.307E-05 | 3.006E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.674E-05 | 3.501E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phosphatidic acid metabolic process | GO:0046473 |  | 2.365E-09 | 5.776E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.142E-08 | 6.281E-05 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 8.553E-07 | 6.965E-04 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 1.813E-06 | 1.107E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.528E-06 | 1.724E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 4.001E-06 | 1.629E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 4.001E-06 | 1.396E-03 |
|---|
| JAK-STAT cascade | GO:0007259 |  | 4.016E-06 | 1.226E-03 |
|---|
| short-chain fatty acid metabolic process | GO:0046459 |  | 5.998E-06 | 1.628E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 5.998E-06 | 1.465E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.998E-06 | 1.332E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.22E-08 | 1.978E-04 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 9.783E-07 | 1.177E-03 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 2.181E-06 | 1.749E-03 |
|---|
| JAK-STAT cascade | GO:0007259 |  | 4.394E-06 | 2.643E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.032E-06 | 2.421E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.087E-06 | 2.04E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 5.087E-06 | 1.748E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 5.087E-06 | 1.53E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.087E-06 | 1.36E-03 |
|---|
| short-chain fatty acid metabolic process | GO:0046459 |  | 7.627E-06 | 1.835E-03 |
|---|
| aging | GO:0007568 |  | 8.659E-06 | 1.894E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phosphatidic acid metabolic process | GO:0046473 |  | 2.365E-09 | 5.776E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.142E-08 | 6.281E-05 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 8.553E-07 | 6.965E-04 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 1.813E-06 | 1.107E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.528E-06 | 1.724E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 4.001E-06 | 1.629E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 4.001E-06 | 1.396E-03 |
|---|
| JAK-STAT cascade | GO:0007259 |  | 4.016E-06 | 1.226E-03 |
|---|
| short-chain fatty acid metabolic process | GO:0046459 |  | 5.998E-06 | 1.628E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 5.998E-06 | 1.465E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.998E-06 | 1.332E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.22E-08 | 1.978E-04 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 9.783E-07 | 1.177E-03 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 2.181E-06 | 1.749E-03 |
|---|
| JAK-STAT cascade | GO:0007259 |  | 4.394E-06 | 2.643E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.032E-06 | 2.421E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.087E-06 | 2.04E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 5.087E-06 | 1.748E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 5.087E-06 | 1.53E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.087E-06 | 1.36E-03 |
|---|
| short-chain fatty acid metabolic process | GO:0046459 |  | 7.627E-06 | 1.835E-03 |
|---|
| aging | GO:0007568 |  | 8.659E-06 | 1.894E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.427E-08 | 8.667E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.366E-06 | 6.009E-03 |
|---|
| phospholipid:diacylglycerol acyltransferase activity | GO:0046027 |  | 4.142E-06 | 4.93E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 4.142E-06 | 3.697E-03 |
|---|
| advanced glycation end-product receptor activity | GO:0050785 |  | 4.142E-06 | 2.958E-03 |
|---|
| phosphopantetheine binding | GO:0031177 |  | 4.142E-06 | 2.465E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 4.142E-06 | 2.113E-03 |
|---|
| short-chain fatty acid metabolic process | GO:0046459 |  | 4.142E-06 | 1.849E-03 |
|---|
| DNA topoisomerase activity | GO:0003916 |  | 4.142E-06 | 1.643E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.21E-06 | 2.218E-03 |
|---|
| DNA topological change | GO:0006265 |  | 6.21E-06 | 2.016E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.38E-08 | 8.681E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.448E-06 | 4.465E-03 |
|---|
| phospholipid:diacylglycerol acyltransferase activity | GO:0046027 |  | 3.763E-06 | 4.576E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 3.763E-06 | 3.432E-03 |
|---|
| acyl carrier activity | GO:0000036 |  | 3.763E-06 | 2.745E-03 |
|---|
| short-chain fatty acid metabolic process | GO:0046459 |  | 3.763E-06 | 2.288E-03 |
|---|
| phosphatidic acid metabolic process | GO:0046473 |  | 3.763E-06 | 1.961E-03 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 3.763E-06 | 1.716E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 3.763E-06 | 1.525E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 5.642E-06 | 2.058E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 5.642E-06 | 1.871E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.22E-08 | 1.978E-04 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 9.783E-07 | 1.177E-03 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 2.181E-06 | 1.749E-03 |
|---|
| JAK-STAT cascade | GO:0007259 |  | 4.394E-06 | 2.643E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.032E-06 | 2.421E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.087E-06 | 2.04E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 5.087E-06 | 1.748E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 5.087E-06 | 1.53E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.087E-06 | 1.36E-03 |
|---|
| short-chain fatty acid metabolic process | GO:0046459 |  | 7.627E-06 | 1.835E-03 |
|---|
| aging | GO:0007568 |  | 8.659E-06 | 1.894E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.411E-07 | 5.544E-04 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 2.785E-06 | 3.203E-03 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 6.376E-06 | 4.888E-03 |
|---|
| JAK-STAT cascade | GO:0007259 |  | 1.048E-05 | 6.028E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.117E-05 | 5.138E-03 |
|---|
| short-chain fatty acid metabolic process | GO:0046459 |  | 1.117E-05 | 4.282E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.117E-05 | 3.67E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.117E-05 | 3.211E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.117E-05 | 2.854E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.307E-05 | 3.006E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.674E-05 | 3.501E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.411E-07 | 5.544E-04 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 2.785E-06 | 3.203E-03 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 6.376E-06 | 4.888E-03 |
|---|
| JAK-STAT cascade | GO:0007259 |  | 1.048E-05 | 6.028E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.117E-05 | 5.138E-03 |
|---|
| short-chain fatty acid metabolic process | GO:0046459 |  | 1.117E-05 | 4.282E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.117E-05 | 3.67E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.117E-05 | 3.211E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.117E-05 | 2.854E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.307E-05 | 3.006E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.674E-05 | 3.501E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| carboxylic acid biosynthetic process | GO:0046394 |  | 1.192E-06 | 2.196E-03 |
|---|
| fatty acid biosynthetic process | GO:0006633 |  | 1.382E-05 | 0.01273508 |
|---|
| acyl-CoA metabolic process | GO:0006637 |  | 1.518E-05 | 9.327E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 1.518E-05 | 6.995E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 2.276E-05 | 8.389E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.276E-05 | 6.99E-03 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 2.276E-05 | 5.992E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 2.276E-05 | 5.243E-03 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 3.184E-05 | 6.52E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 3.184E-05 | 5.868E-03 |
|---|
| regulation of stress fiber formation | GO:0051492 |  | 4.242E-05 | 7.108E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.411E-07 | 5.544E-04 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 2.785E-06 | 3.203E-03 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 6.376E-06 | 4.888E-03 |
|---|
| JAK-STAT cascade | GO:0007259 |  | 1.048E-05 | 6.028E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.117E-05 | 5.138E-03 |
|---|
| short-chain fatty acid metabolic process | GO:0046459 |  | 1.117E-05 | 4.282E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.117E-05 | 3.67E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.117E-05 | 3.211E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.117E-05 | 2.854E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.307E-05 | 3.006E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.674E-05 | 3.501E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.172E-07 | 7.528E-04 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 3.78E-06 | 4.485E-03 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 8.908E-06 | 7.047E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.166E-05 | 6.916E-03 |
|---|
| short-chain fatty acid metabolic process | GO:0046459 |  | 1.166E-05 | 5.533E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.166E-05 | 4.611E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.166E-05 | 3.952E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.511E-05 | 4.482E-03 |
|---|
| JAK-STAT cascade | GO:0007259 |  | 1.511E-05 | 3.984E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.747E-05 | 4.147E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.445E-05 | 5.274E-03 |
|---|



