Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 5424-6-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1948 | 2.524e-02 | 2.134e-02 | 1.777e-01 |
|---|
| IPC-NIBC-129 | 0.2648 | 3.214e-02 | 4.712e-02 | 3.313e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2046 | 1.149e-02 | 1.889e-02 | 2.938e-01 |
|---|
| Loi_GPL570 | 0.2064 | 1.076e-01 | 1.198e-01 | 3.887e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1595 | 1.000e-06 | 1.200e-05 | 1.000e-06 |
|---|
| Parker_GPL1390 | 0.2334 | 3.330e-02 | 4.215e-02 | 3.130e-01 |
|---|
| Parker_GPL887 | 0.0128 | 3.712e-01 | 3.901e-01 | 4.064e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3423 | 9.753e-01 | 9.689e-01 | 1.000e+00 |
|---|
| Schmidt | 0.2712 | 1.356e-02 | 1.526e-02 | 1.785e-01 |
|---|
| Sotiriou | 0.2596 | 2.889e-02 | 2.285e-02 | 3.355e-01 |
|---|
| Van-De-Vijver | 0.2890 | 1.217e-03 | 1.752e-03 | 6.691e-02 |
|---|
| Zhang | 0.2943 | 1.353e-01 | 8.894e-02 | 2.826e-01 |
|---|
| Zhou | 0.4093 | 7.814e-02 | 6.766e-02 | 4.399e-01 |
|---|
Expression data for subnetwork 5424-6-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
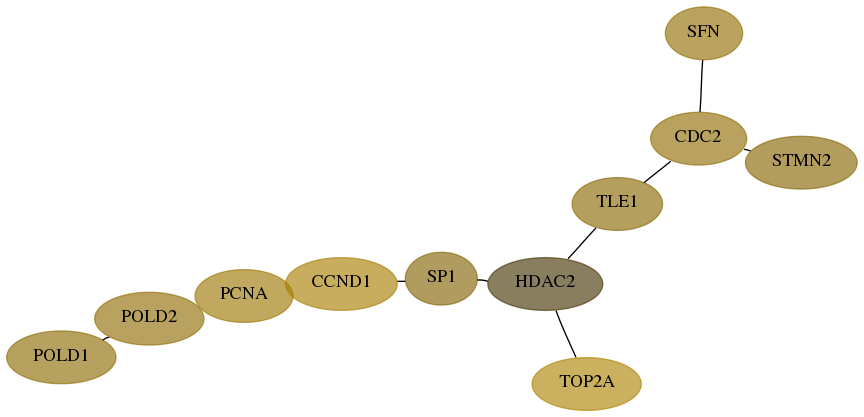
Score for each gene in subnetwork 5424-6-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| pold2 |   | 1 | 116 | 279 | 280 | 0.073 | 0.180 | 0.204 | 0.131 | 0.150 | 0.084 | 0.376 | 0.186 | 0.146 | 0.126 | 0.174 | 0.072 | 0.256 |
|---|
| pcna |   | 15 | 11 | 110 | 85 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.119 | 0.104 | 0.256 | 0.151 | 0.059 | 0.201 | 0.123 | 0.233 |
|---|
| stmn2 |   | 29 | 9 | 34 | 27 | 0.057 | 0.106 | 0.005 | 0.052 | 0.110 | 0.071 | -0.099 | 0.006 | 0.034 | 0.103 | 0.008 | 0.113 | -0.039 |
|---|
| tle1 |   | 47 | 6 | 1 | 3 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | 0.170 | 0.068 | 0.025 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| sp1 |   | 8 | 26 | 71 | 57 | 0.054 | 0.186 | 0.147 | 0.242 | 0.125 | 0.033 | -0.370 | 0.202 | 0.284 | 0.227 | 0.304 | 0.091 | -0.068 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| hdac2 |   | 3 | 62 | 146 | 135 | 0.012 | 0.245 | 0.030 | 0.077 | 0.070 | 0.307 | -0.204 | 0.183 | 0.222 | -0.058 | 0.038 | 0.133 | 0.181 |
|---|
| sfn |   | 61 | 3 | 1 | 2 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.158 | 0.079 | 0.181 |
|---|
| pold1 |   | 1 | 116 | 279 | 280 | 0.069 | 0.107 | 0.160 | -0.033 | 0.102 | 0.109 | 0.104 | 0.121 | 0.144 | 0.148 | 0.230 | 0.006 | 0.410 |
|---|
| top2a |   | 15 | 11 | 163 | 134 | 0.147 | 0.183 | 0.185 | 0.100 | 0.206 | 0.014 | 0.035 | 0.309 | 0.261 | 0.183 | 0.147 | 0.234 | 0.273 |
|---|
GO Enrichment output for subnetwork 5424-6-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.247E-09 | 5.167E-06 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 3.255E-07 | 3.743E-04 |
|---|
| base-excision repair | GO:0006284 |  | 2.552E-06 | 1.957E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.241E-05 | 7.135E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.241E-05 | 5.708E-03 |
|---|
| maintenance of fidelity during DNA-dependent DNA replication | GO:0045005 |  | 1.241E-05 | 4.757E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.762E-05 | 5.79E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.86E-05 | 5.348E-03 |
|---|
| response to UV | GO:0009411 |  | 1.884E-05 | 4.814E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.575E-05 | 5.922E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.602E-05 | 5.441E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.992E-10 | 4.868E-07 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 4.253E-08 | 5.195E-05 |
|---|
| base-excision repair | GO:0006284 |  | 6.674E-07 | 5.435E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.496E-06 | 2.135E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 3.653E-06 | 1.785E-03 |
|---|
| maintenance of fidelity during DNA-dependent DNA replication | GO:0045005 |  | 3.653E-06 | 1.487E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 3.653E-06 | 1.275E-03 |
|---|
| response to UV | GO:0009411 |  | 5.287E-06 | 1.615E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 5.477E-06 | 1.487E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 5.477E-06 | 1.338E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.892E-06 | 1.309E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.862E-10 | 9.293E-07 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 7.647E-08 | 9.199E-05 |
|---|
| base-excision repair | GO:0006284 |  | 9.783E-07 | 7.846E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 5.087E-06 | 3.06E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 5.087E-06 | 2.448E-03 |
|---|
| maintenance of fidelity during DNA-dependent DNA replication | GO:0045005 |  | 5.087E-06 | 2.04E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 5.087E-06 | 1.748E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.728E-06 | 1.723E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 7.627E-06 | 2.039E-03 |
|---|
| response to UV | GO:0009411 |  | 7.739E-06 | 1.862E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.648E-06 | 2.11E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.992E-10 | 4.868E-07 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 4.253E-08 | 5.195E-05 |
|---|
| base-excision repair | GO:0006284 |  | 6.674E-07 | 5.435E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.496E-06 | 2.135E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 3.653E-06 | 1.785E-03 |
|---|
| maintenance of fidelity during DNA-dependent DNA replication | GO:0045005 |  | 3.653E-06 | 1.487E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 3.653E-06 | 1.275E-03 |
|---|
| response to UV | GO:0009411 |  | 5.287E-06 | 1.615E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 5.477E-06 | 1.487E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 5.477E-06 | 1.338E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.892E-06 | 1.309E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.862E-10 | 9.293E-07 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 7.647E-08 | 9.199E-05 |
|---|
| base-excision repair | GO:0006284 |  | 9.783E-07 | 7.846E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 5.087E-06 | 3.06E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 5.087E-06 | 2.448E-03 |
|---|
| maintenance of fidelity during DNA-dependent DNA replication | GO:0045005 |  | 5.087E-06 | 2.04E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 5.087E-06 | 1.748E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.728E-06 | 1.723E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 7.627E-06 | 2.039E-03 |
|---|
| response to UV | GO:0009411 |  | 7.739E-06 | 1.862E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.648E-06 | 2.11E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.535E-10 | 9.054E-07 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.259E-08 | 2.247E-05 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 3.589E-08 | 4.272E-05 |
|---|
| base-excision repair | GO:0006284 |  | 2.975E-07 | 2.656E-04 |
|---|
| DNA-directed DNA polymerase activity | GO:0003887 |  | 4.523E-07 | 3.23E-04 |
|---|
| DNA polymerase activity | GO:0034061 |  | 7.31E-07 | 4.351E-04 |
|---|
| replication fork | GO:0005657 |  | 1.002E-06 | 5.111E-04 |
|---|
| response to UV | GO:0009411 |  | 4.081E-06 | 1.821E-03 |
|---|
| embryonic camera-type eye development | GO:0031076 |  | 4.142E-06 | 1.643E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 4.142E-06 | 1.479E-03 |
|---|
| MutLalpha complex binding | GO:0032405 |  | 4.142E-06 | 1.344E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.532E-10 | 9.237E-07 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.462E-08 | 2.667E-05 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 2.911E-08 | 3.539E-05 |
|---|
| DNA-directed DNA polymerase activity | GO:0003887 |  | 5.964E-07 | 5.44E-04 |
|---|
| replication fork | GO:0005657 |  | 9.46E-07 | 6.902E-04 |
|---|
| DNA polymerase activity | GO:0034061 |  | 9.46E-07 | 5.752E-04 |
|---|
| response to UV | GO:0009411 |  | 3.65E-06 | 1.902E-03 |
|---|
| enucleate erythrocyte differentiation | GO:0043353 |  | 4.447E-06 | 2.028E-03 |
|---|
| S phase | GO:0051320 |  | 4.447E-06 | 1.802E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 4.447E-06 | 1.622E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 4.447E-06 | 1.475E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.862E-10 | 9.293E-07 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 7.647E-08 | 9.199E-05 |
|---|
| base-excision repair | GO:0006284 |  | 9.783E-07 | 7.846E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 5.087E-06 | 3.06E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 5.087E-06 | 2.448E-03 |
|---|
| maintenance of fidelity during DNA-dependent DNA replication | GO:0045005 |  | 5.087E-06 | 2.04E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 5.087E-06 | 1.748E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.728E-06 | 1.723E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 7.627E-06 | 2.039E-03 |
|---|
| response to UV | GO:0009411 |  | 7.739E-06 | 1.862E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.648E-06 | 2.11E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.247E-09 | 5.167E-06 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 3.255E-07 | 3.743E-04 |
|---|
| base-excision repair | GO:0006284 |  | 2.552E-06 | 1.957E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.241E-05 | 7.135E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.241E-05 | 5.708E-03 |
|---|
| maintenance of fidelity during DNA-dependent DNA replication | GO:0045005 |  | 1.241E-05 | 4.757E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.762E-05 | 5.79E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.86E-05 | 5.348E-03 |
|---|
| response to UV | GO:0009411 |  | 1.884E-05 | 4.814E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.575E-05 | 5.922E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.602E-05 | 5.441E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.247E-09 | 5.167E-06 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 3.255E-07 | 3.743E-04 |
|---|
| base-excision repair | GO:0006284 |  | 2.552E-06 | 1.957E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.241E-05 | 7.135E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.241E-05 | 5.708E-03 |
|---|
| maintenance of fidelity during DNA-dependent DNA replication | GO:0045005 |  | 1.241E-05 | 4.757E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.762E-05 | 5.79E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.86E-05 | 5.348E-03 |
|---|
| response to UV | GO:0009411 |  | 1.884E-05 | 4.814E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.575E-05 | 5.922E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.602E-05 | 5.441E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 1.6E-09 | 2.948E-06 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.807E-07 | 1.665E-04 |
|---|
| base-excision repair | GO:0006284 |  | 7.984E-07 | 4.905E-04 |
|---|
| DNA replication | GO:0006260 |  | 5.661E-06 | 2.608E-03 |
|---|
| chromosome segregation | GO:0007059 |  | 1.817E-05 | 6.696E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 1.897E-05 | 5.828E-03 |
|---|
| positive regulation of viral reproduction | GO:0048524 |  | 1.897E-05 | 4.995E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 2.844E-05 | 6.551E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 3.039E-05 | 6.224E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 3.978E-05 | 7.331E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 5.3E-05 | 8.88E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.247E-09 | 5.167E-06 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 3.255E-07 | 3.743E-04 |
|---|
| base-excision repair | GO:0006284 |  | 2.552E-06 | 1.957E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.241E-05 | 7.135E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.241E-05 | 5.708E-03 |
|---|
| maintenance of fidelity during DNA-dependent DNA replication | GO:0045005 |  | 1.241E-05 | 4.757E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.762E-05 | 5.79E-03 |
|---|
| DNA topological change | GO:0006265 |  | 1.86E-05 | 5.348E-03 |
|---|
| response to UV | GO:0009411 |  | 1.884E-05 | 4.814E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.575E-05 | 5.922E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.602E-05 | 5.441E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 2.647E-09 | 6.28E-06 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 3.424E-07 | 4.062E-04 |
|---|
| base-excision repair | GO:0006284 |  | 3.005E-06 | 2.377E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.166E-05 | 6.916E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.166E-05 | 5.533E-03 |
|---|
| maintenance of fidelity during DNA-dependent DNA replication | GO:0045005 |  | 1.166E-05 | 4.611E-03 |
|---|
| definitive hemopoiesis | GO:0060216 |  | 1.166E-05 | 3.952E-03 |
|---|
| embryonic camera-type eye development | GO:0031076 |  | 1.747E-05 | 5.183E-03 |
|---|
| response to UV | GO:0009411 |  | 1.846E-05 | 4.867E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.445E-05 | 5.802E-03 |
|---|
| DNA topological change | GO:0006265 |  | 2.445E-05 | 5.274E-03 |
|---|



