Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 523-6-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1791 | 1.029e-02 | 7.734e-03 | 8.916e-02 |
|---|
| IPC-NIBC-129 | 0.2483 | 6.446e-02 | 9.166e-02 | 5.185e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2485 | 8.480e-03 | 1.431e-02 | 2.486e-01 |
|---|
| Loi_GPL570 | 0.2570 | 5.302e-02 | 6.023e-02 | 2.289e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1753 | 3.000e-06 | 3.300e-05 | 9.000e-06 |
|---|
| Parker_GPL1390 | 0.2474 | 8.309e-02 | 1.007e-01 | 5.131e-01 |
|---|
| Parker_GPL887 | 0.0749 | 4.083e-01 | 4.274e-01 | 4.521e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3542 | 6.675e-01 | 6.067e-01 | 9.926e-01 |
|---|
| Schmidt | 0.2073 | 1.733e-02 | 1.985e-02 | 2.121e-01 |
|---|
| Sotiriou | 0.3099 | 1.260e-02 | 9.703e-03 | 2.148e-01 |
|---|
| Van-De-Vijver | 0.2383 | 1.136e-03 | 1.635e-03 | 6.391e-02 |
|---|
| Zhang | 0.2794 | 1.065e-01 | 6.580e-02 | 2.315e-01 |
|---|
| Zhou | 0.4132 | 7.007e-02 | 5.875e-02 | 4.069e-01 |
|---|
Expression data for subnetwork 523-6-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
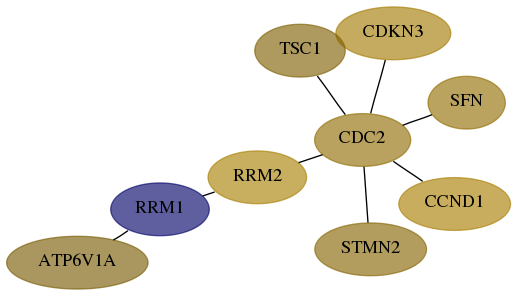
Score for each gene in subnetwork 523-6-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| cdkn3 |   | 53 | 5 | 29 | 22 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.166 | -0.009 | 0.323 | 0.186 | 0.198 | 0.201 | 0.271 | 0.372 |
|---|
| rrm2 |   | 5 | 40 | 163 | 143 | 0.131 | 0.207 | 0.227 | 0.187 | 0.161 | 0.178 | 0.137 | 0.303 | 0.190 | 0.219 | undef | 0.129 | 0.302 |
|---|
| stmn2 |   | 29 | 9 | 34 | 27 | 0.057 | 0.106 | 0.005 | 0.052 | 0.110 | 0.071 | -0.099 | 0.006 | 0.034 | 0.103 | 0.008 | 0.113 | -0.039 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| rrm1 |   | 1 | 116 | 225 | 228 | -0.029 | 0.117 | 0.065 | 0.106 | 0.111 | 0.148 | -0.018 | 0.185 | 0.172 | 0.140 | 0.133 | 0.135 | 0.263 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| sfn |   | 61 | 3 | 1 | 2 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.158 | 0.079 | 0.181 |
|---|
| atp6v1a |   | 1 | 116 | 225 | 228 | 0.041 | 0.138 | 0.073 | 0.044 | 0.085 | 0.155 | -0.204 | 0.067 | 0.041 | 0.038 | 0.053 | 0.028 | 0.042 |
|---|
GO Enrichment output for subnetwork 523-6-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 5.258E-12 | 1.209E-08 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.082E-09 | 2.394E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.785E-08 | 1.368E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.563E-08 | 4.924E-05 |
|---|
| protein oligomerization | GO:0051259 |  | 2.304E-07 | 1.06E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.178E-06 | 2.368E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.861E-06 | 2.254E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 6.861E-06 | 1.973E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 3.679E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.439E-05 | 3.311E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 1.918E-05 | 4.011E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.616E-09 | 6.391E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.066E-08 | 1.302E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.318E-07 | 1.073E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.123E-06 | 6.856E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.898E-06 | 9.275E-04 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 2.847E-06 | 1.159E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 5.311E-06 | 1.853E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 5.311E-06 | 1.622E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 5.795E-06 | 1.573E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 6.826E-06 | 1.668E-03 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.826E-06 | 1.516E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.857E-12 | 4.468E-09 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.862E-10 | 4.646E-07 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.031E-08 | 8.271E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.062E-08 | 1.241E-05 |
|---|
| protein oligomerization | GO:0051259 |  | 7.398E-08 | 3.56E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.841E-06 | 7.384E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.644E-06 | 9.086E-04 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 2.644E-06 | 7.951E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.395E-06 | 1.977E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 7.395E-06 | 1.779E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 9.505E-06 | 2.079E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.616E-09 | 6.391E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.066E-08 | 1.302E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.318E-07 | 1.073E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.123E-06 | 6.856E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.898E-06 | 9.275E-04 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 2.847E-06 | 1.159E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 5.311E-06 | 1.853E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 5.311E-06 | 1.622E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 5.795E-06 | 1.573E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 6.826E-06 | 1.668E-03 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.826E-06 | 1.516E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.857E-12 | 4.468E-09 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.862E-10 | 4.646E-07 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.031E-08 | 8.271E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.062E-08 | 1.241E-05 |
|---|
| protein oligomerization | GO:0051259 |  | 7.398E-08 | 3.56E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.841E-06 | 7.384E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.644E-06 | 9.086E-04 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 2.644E-06 | 7.951E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.395E-06 | 1.977E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 7.395E-06 | 1.779E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 9.505E-06 | 2.079E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| oxidoreductase activity. acting on CH or CH2 groups | GO:0016725 |  | 3.674E-09 | 1.312E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.674E-09 | 6.559E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.131E-09 | 9.679E-06 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 8.541E-08 | 7.625E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.578E-06 | 1.127E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.578E-06 | 1.534E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.865E-06 | 1.972E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 4.109E-06 | 1.834E-03 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 4.31E-06 | 1.71E-03 |
|---|
| proton-transporting V-type ATPase. V1 domain | GO:0033180 |  | 5.41E-06 | 1.932E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 5.41E-06 | 1.756E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.194E-09 | 8.002E-06 |
|---|
| oxidoreductase activity. acting on CH or CH2 groups | GO:0016725 |  | 3.838E-09 | 7E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.014E-08 | 1.233E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.059E-07 | 9.657E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.34E-06 | 9.773E-04 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 2.566E-06 | 1.56E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 2.566E-06 | 1.337E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.848E-06 | 1.755E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 3.848E-06 | 1.56E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.848E-06 | 1.404E-03 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 4.935E-06 | 1.637E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.857E-12 | 4.468E-09 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.862E-10 | 4.646E-07 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.031E-08 | 8.271E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.062E-08 | 1.241E-05 |
|---|
| protein oligomerization | GO:0051259 |  | 7.398E-08 | 3.56E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.841E-06 | 7.384E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.644E-06 | 9.086E-04 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 2.644E-06 | 7.951E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.395E-06 | 1.977E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 7.395E-06 | 1.779E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 9.505E-06 | 2.079E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 5.258E-12 | 1.209E-08 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.082E-09 | 2.394E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.785E-08 | 1.368E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.563E-08 | 4.924E-05 |
|---|
| protein oligomerization | GO:0051259 |  | 2.304E-07 | 1.06E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.178E-06 | 2.368E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.861E-06 | 2.254E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 6.861E-06 | 1.973E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 3.679E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.439E-05 | 3.311E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 1.918E-05 | 4.011E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 5.258E-12 | 1.209E-08 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.082E-09 | 2.394E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.785E-08 | 1.368E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.563E-08 | 4.924E-05 |
|---|
| protein oligomerization | GO:0051259 |  | 2.304E-07 | 1.06E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.178E-06 | 2.368E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.861E-06 | 2.254E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 6.861E-06 | 1.973E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 3.679E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.439E-05 | 3.311E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 1.918E-05 | 4.011E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell growth | GO:0030307 |  | 9.493E-06 | 0.01749647 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 9.493E-06 | 8.748E-03 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 9.493E-06 | 5.832E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 9.493E-06 | 4.374E-03 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 1.329E-05 | 4.897E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 1.329E-05 | 4.081E-03 |
|---|
| regulation of stress fiber formation | GO:0051492 |  | 1.771E-05 | 4.662E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.771E-05 | 4.079E-03 |
|---|
| positive regulation of cell size | GO:0045793 |  | 2.276E-05 | 4.66E-03 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 2.276E-05 | 4.194E-03 |
|---|
| regulation of dephosphorylation | GO:0035303 |  | 2.276E-05 | 3.813E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 5.258E-12 | 1.209E-08 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.082E-09 | 2.394E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.785E-08 | 1.368E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.563E-08 | 4.924E-05 |
|---|
| protein oligomerization | GO:0051259 |  | 2.304E-07 | 1.06E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.178E-06 | 2.368E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.861E-06 | 2.254E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 6.861E-06 | 1.973E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 3.679E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.439E-05 | 3.311E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 1.918E-05 | 4.011E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.641E-11 | 3.893E-08 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.4E-09 | 4.034E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 3.476E-08 | 2.75E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.338E-07 | 7.935E-05 |
|---|
| protein oligomerization | GO:0051259 |  | 3.268E-07 | 1.551E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.724E-06 | 3.055E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.055E-06 | 2.731E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.158E-05 | 3.435E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.62E-05 | 4.272E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 2.159E-05 | 5.124E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.159E-05 | 4.658E-03 |
|---|



