Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 4291-5-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2026 | 1.408e-02 | 1.104e-02 | 1.141e-01 |
|---|
| IPC-NIBC-129 | 0.2295 | 1.785e-02 | 2.670e-02 | 2.126e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2332 | 5.347e-03 | 9.389e-03 | 1.905e-01 |
|---|
| Loi_GPL570 | 0.2423 | 6.530e-02 | 7.377e-02 | 2.692e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1592 | 1.200e-05 | 1.050e-04 | 8.600e-05 |
|---|
| Parker_GPL1390 | 0.2686 | 3.308e-02 | 4.189e-02 | 3.118e-01 |
|---|
| Parker_GPL887 | 0.2396 | 3.540e-01 | 3.727e-01 | 3.851e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3094 | 1.354e-01 | 8.925e-02 | 6.306e-01 |
|---|
| Schmidt | 0.2717 | 1.653e-02 | 1.887e-02 | 2.052e-01 |
|---|
| Sotiriou | 0.4144 | 2.165e-03 | 1.588e-03 | 7.483e-02 |
|---|
| Van-De-Vijver | 0.3314 | 7.320e-04 | 1.054e-03 | 4.757e-02 |
|---|
| Zhang | 0.2823 | 1.360e-01 | 8.947e-02 | 2.837e-01 |
|---|
| Zhou | 0.4377 | 1.051e-01 | 9.890e-02 | 5.358e-01 |
|---|
Expression data for subnetwork 4291-5-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
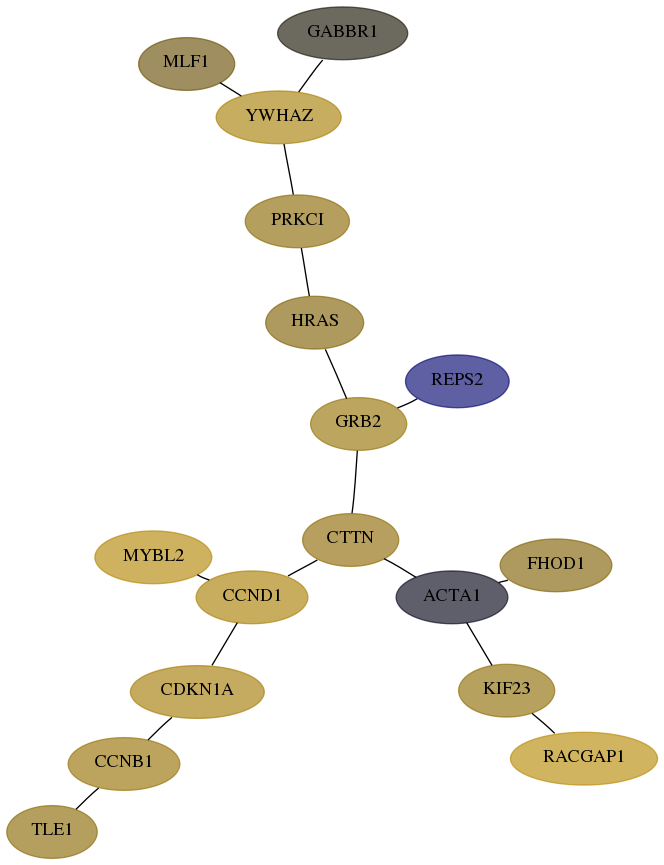
Score for each gene in subnetwork 4291-5-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| cttn |   | 10 | 20 | 1 | 6 | 0.067 | 0.065 | 0.123 | 0.238 | 0.126 | 0.263 | -0.172 | 0.216 | 0.069 | 0.115 | undef | 0.025 | 0.279 |
|---|
| cdkn1a |   | 44 | 7 | 1 | 4 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | undef | -0.201 | 0.069 |
|---|
| tle1 |   | 47 | 6 | 1 | 3 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | 0.170 | 0.068 | 0.025 |
|---|
| ccnb1 |   | 42 | 8 | 56 | 39 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | undef | 0.167 | 0.398 |
|---|
| prkci |   | 14 | 15 | 31 | 26 | 0.065 | 0.104 | 0.012 | 0.170 | 0.112 | 0.088 | -0.103 | -0.170 | 0.196 | 0.128 | 0.049 | 0.124 | 0.149 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| hras |   | 14 | 15 | 42 | 34 | 0.049 | -0.030 | 0.094 | 0.210 | 0.160 | 0.195 | 0.022 | 0.245 | 0.086 | 0.208 | 0.180 | 0.003 | 0.197 |
|---|
| gabbr1 |   | 7 | 30 | 56 | 49 | 0.003 | -0.016 | -0.022 | -0.051 | 0.082 | 0.119 | 0.584 | -0.082 | 0.049 | 0.057 | 0.125 | 0.096 | 0.116 |
|---|
| mlf1 |   | 7 | 30 | 56 | 49 | 0.027 | 0.224 | 0.053 | 0.047 | 0.169 | 0.128 | 0.021 | -0.028 | 0.106 | 0.176 | undef | 0.098 | 0.141 |
|---|
| racgap1 |   | 1 | 116 | 56 | 70 | 0.181 | 0.240 | 0.218 | 0.126 | 0.242 | 0.125 | 0.092 | 0.313 | 0.238 | 0.255 | 0.253 | 0.234 | 0.356 |
|---|
| mybl2 |   | 9 | 22 | 56 | 48 | 0.162 | 0.237 | 0.166 | 0.079 | 0.124 | 0.184 | 0.333 | 0.275 | 0.202 | 0.182 | 0.289 | 0.074 | 0.245 |
|---|
| reps2 |   | 3 | 62 | 56 | 56 | -0.033 | -0.139 | 0.057 | 0.122 | 0.113 | -0.154 | 0.105 | -0.157 | 0.079 | 0.124 | 0.052 | 0.303 | 0.220 |
|---|
| kif23 |   | 1 | 116 | 56 | 70 | 0.068 | 0.197 | 0.069 | 0.192 | 0.117 | 0.180 | 0.112 | 0.311 | 0.230 | 0.130 | undef | 0.070 | 0.390 |
|---|
| fhod1 |   | 1 | 116 | 56 | 70 | 0.050 | 0.216 | 0.126 | -0.110 | 0.073 | 0.134 | -0.082 | 0.094 | -0.080 | -0.013 | 0.112 | 0.075 | 0.207 |
|---|
| ywhaz |   | 17 | 10 | 29 | 25 | 0.124 | 0.089 | 0.087 | 0.200 | 0.141 | 0.110 | 0.228 | 0.199 | 0.056 | 0.123 | undef | 0.103 | 0.213 |
|---|
| acta1 |   | 9 | 22 | 1 | 7 | -0.003 | 0.098 | -0.008 | -0.030 | 0.119 | 0.174 | 0.376 | 0.024 | 0.182 | 0.214 | 0.100 | 0.048 | 0.201 |
|---|
| grb2 |   | 9 | 22 | 1 | 7 | 0.084 | -0.013 | 0.086 | -0.031 | 0.162 | -0.038 | 0.003 | 0.065 | 0.158 | 0.127 | 0.149 | 0.194 | -0.005 |
|---|
GO Enrichment output for subnetwork 4291-5-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 4.55E-08 | 1.046E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.455E-07 | 8.573E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.515E-06 | 1.929E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.097E-06 | 2.931E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 5.94E-06 | 2.732E-03 |
|---|
| sperm motility | GO:0030317 |  | 9.008E-06 | 3.453E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.154E-05 | 3.793E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.357E-05 | 3.901E-03 |
|---|
| interphase | GO:0051325 |  | 1.486E-05 | 3.798E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.617E-05 | 3.719E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 2.097E-05 | 4.385E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.037E-08 | 2.533E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.948E-07 | 3.601E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 5.76E-07 | 4.691E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 1.363E-06 | 8.326E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.351E-06 | 1.149E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 2.655E-06 | 1.081E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.655E-06 | 9.267E-04 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 3.341E-06 | 1.02E-03 |
|---|
| sperm motility | GO:0030317 |  | 3.341E-06 | 9.069E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.346E-06 | 8.174E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 5.044E-06 | 1.12E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.704E-08 | 4.1E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.84E-07 | 5.823E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 9.453E-07 | 7.582E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 1.918E-06 | 1.154E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 3.394E-06 | 1.633E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.853E-06 | 1.545E-03 |
|---|
| sperm motility | GO:0030317 |  | 3.853E-06 | 1.324E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 3.853E-06 | 1.159E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 4.892E-06 | 1.308E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.664E-06 | 1.363E-03 |
|---|
| interphase | GO:0051325 |  | 6.41E-06 | 1.402E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.037E-08 | 2.533E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.948E-07 | 3.601E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 5.76E-07 | 4.691E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 1.363E-06 | 8.326E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.351E-06 | 1.149E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 2.655E-06 | 1.081E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.655E-06 | 9.267E-04 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 3.341E-06 | 1.02E-03 |
|---|
| sperm motility | GO:0030317 |  | 3.341E-06 | 9.069E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.346E-06 | 8.174E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 5.044E-06 | 1.12E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.704E-08 | 4.1E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.84E-07 | 5.823E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 9.453E-07 | 7.582E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 1.918E-06 | 1.154E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 3.394E-06 | 1.633E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.853E-06 | 1.545E-03 |
|---|
| sperm motility | GO:0030317 |  | 3.853E-06 | 1.324E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 3.853E-06 | 1.159E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 4.892E-06 | 1.308E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.664E-06 | 1.363E-03 |
|---|
| interphase | GO:0051325 |  | 6.41E-06 | 1.402E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 8.541E-08 | 3.05E-04 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 2.511E-07 | 4.483E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.03E-06 | 1.226E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.569E-06 | 2.294E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 3.463E-06 | 2.473E-03 |
|---|
| actomyosin structure organization | GO:0031032 |  | 4.397E-06 | 2.617E-03 |
|---|
| striated muscle cell differentiation | GO:0051146 |  | 6.736E-06 | 3.436E-03 |
|---|
| skeletal myofibril assembly | GO:0014866 |  | 1.407E-05 | 6.279E-03 |
|---|
| alpha-tubulin binding | GO:0043014 |  | 1.407E-05 | 5.581E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.108E-05 | 7.529E-03 |
|---|
| insulin receptor substrate binding | GO:0043560 |  | 2.108E-05 | 6.844E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 4.948E-08 | 1.805E-04 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 3.204E-07 | 5.844E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.923E-07 | 5.986E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.2E-06 | 1.094E-03 |
|---|
| interphase | GO:0051325 |  | 1.324E-06 | 9.657E-04 |
|---|
| actomyosin structure organization | GO:0031032 |  | 1.349E-06 | 8.199E-04 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 2.271E-06 | 1.183E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 2.794E-06 | 1.274E-03 |
|---|
| striated muscle cell differentiation | GO:0051146 |  | 3.185E-06 | 1.291E-03 |
|---|
| muscle thin filament assembly | GO:0030240 |  | 9.742E-06 | 3.554E-03 |
|---|
| hemopoietic progenitor cell differentiation | GO:0002244 |  | 9.742E-06 | 3.231E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.704E-08 | 4.1E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.84E-07 | 5.823E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 9.453E-07 | 7.582E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 1.918E-06 | 1.154E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 3.394E-06 | 1.633E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.853E-06 | 1.545E-03 |
|---|
| sperm motility | GO:0030317 |  | 3.853E-06 | 1.324E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 3.853E-06 | 1.159E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 4.892E-06 | 1.308E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.664E-06 | 1.363E-03 |
|---|
| interphase | GO:0051325 |  | 6.41E-06 | 1.402E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 4.55E-08 | 1.046E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.455E-07 | 8.573E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.515E-06 | 1.929E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.097E-06 | 2.931E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 5.94E-06 | 2.732E-03 |
|---|
| sperm motility | GO:0030317 |  | 9.008E-06 | 3.453E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.154E-05 | 3.793E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.357E-05 | 3.901E-03 |
|---|
| interphase | GO:0051325 |  | 1.486E-05 | 3.798E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.617E-05 | 3.719E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 2.097E-05 | 4.385E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 4.55E-08 | 1.046E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.455E-07 | 8.573E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.515E-06 | 1.929E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.097E-06 | 2.931E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 5.94E-06 | 2.732E-03 |
|---|
| sperm motility | GO:0030317 |  | 9.008E-06 | 3.453E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.154E-05 | 3.793E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.357E-05 | 3.901E-03 |
|---|
| interphase | GO:0051325 |  | 1.486E-05 | 3.798E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.617E-05 | 3.719E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 2.097E-05 | 4.385E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| actomyosin structure organization | GO:0031032 |  | 4.833E-07 | 8.907E-04 |
|---|
| striated muscle cell differentiation | GO:0051146 |  | 9.972E-07 | 9.189E-04 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.784E-06 | 1.096E-03 |
|---|
| muscle cell differentiation | GO:0042692 |  | 1.293E-05 | 5.956E-03 |
|---|
| regulation of long-term neuronal synaptic plasticity | GO:0048169 |  | 1.897E-05 | 6.993E-03 |
|---|
| establishment or maintenance of epithelial cell apical/basal polarity | GO:0045197 |  | 1.897E-05 | 5.828E-03 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 1.897E-05 | 4.995E-03 |
|---|
| establishment of cell polarity | GO:0030010 |  | 1.897E-05 | 4.371E-03 |
|---|
| neuroblast proliferation | GO:0007405 |  | 2.844E-05 | 5.823E-03 |
|---|
| establishment or maintenance of apical/basal cell polarity | GO:0035088 |  | 2.844E-05 | 5.241E-03 |
|---|
| regulation of synaptic transmission. GABAergic | GO:0032228 |  | 2.844E-05 | 4.764E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 4.55E-08 | 1.046E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 7.455E-07 | 8.573E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.515E-06 | 1.929E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.097E-06 | 2.931E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 5.94E-06 | 2.732E-03 |
|---|
| sperm motility | GO:0030317 |  | 9.008E-06 | 3.453E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.154E-05 | 3.793E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.357E-05 | 3.901E-03 |
|---|
| interphase | GO:0051325 |  | 1.486E-05 | 3.798E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.617E-05 | 3.719E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 2.097E-05 | 4.385E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 3.091E-08 | 7.335E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 6.752E-07 | 8.011E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.076E-06 | 1.642E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.044E-06 | 2.399E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 4.678E-06 | 2.22E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 7.866E-06 | 3.111E-03 |
|---|
| sperm motility | GO:0030317 |  | 7.866E-06 | 2.667E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 8.582E-06 | 2.546E-03 |
|---|
| interphase | GO:0051325 |  | 9.394E-06 | 2.477E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.353E-05 | 3.211E-03 |
|---|
| actomyosin structure organization | GO:0031032 |  | 1.796E-05 | 3.874E-03 |
|---|



