Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 3956-4-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2619 | 1.205e-02 | 9.257e-03 | 1.011e-01 |
|---|
| IPC-NIBC-129 | 0.2536 | 3.498e-02 | 5.110e-02 | 3.514e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2408 | 4.809e-03 | 8.521e-03 | 1.788e-01 |
|---|
| Loi_GPL570 | 0.3286 | 1.888e-02 | 2.201e-02 | 9.660e-02 |
|---|
| Loi_GPL96-GPL97 | 0.1438 | 2.000e-06 | 2.400e-05 | 5.000e-06 |
|---|
| Parker_GPL1390 | 0.2734 | 2.737e-02 | 3.495e-02 | 2.787e-01 |
|---|
| Parker_GPL887 | 0.0896 | 2.444e-01 | 2.612e-01 | 2.502e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3584 | 5.918e-01 | 5.239e-01 | 9.844e-01 |
|---|
| Schmidt | 0.2847 | 4.021e-02 | 4.854e-02 | 3.661e-01 |
|---|
| Sotiriou | 0.3246 | 9.858e-03 | 7.537e-03 | 1.869e-01 |
|---|
| Van-De-Vijver | 0.2829 | 1.625e-03 | 2.338e-03 | 8.089e-02 |
|---|
| Zhang | 0.2280 | 1.709e-01 | 1.192e-01 | 3.417e-01 |
|---|
| Zhou | 0.3932 | 6.743e-02 | 5.589e-02 | 3.957e-01 |
|---|
Expression data for subnetwork 3956-4-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
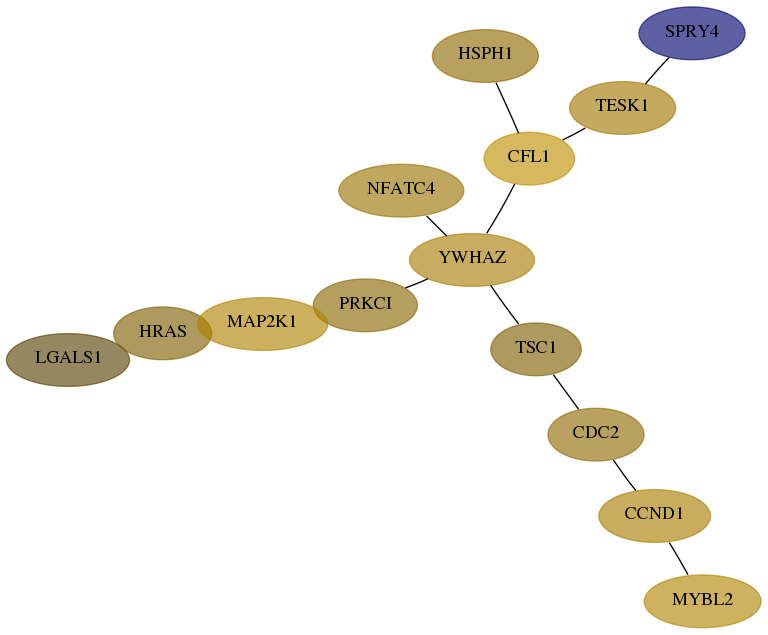
Score for each gene in subnetwork 3956-4-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| nfatc4 |   | 15 | 11 | 1 | 5 | 0.094 | -0.056 | -0.021 | 0.204 | 0.079 | -0.008 | 0.243 | 0.060 | 0.077 | -0.012 | 0.136 | 0.131 | 0.154 |
|---|
| mybl2 |   | 9 | 22 | 56 | 48 | 0.162 | 0.237 | 0.166 | 0.079 | 0.124 | 0.184 | 0.333 | 0.275 | 0.202 | 0.182 | 0.289 | 0.074 | 0.245 |
|---|
| ywhaz |   | 17 | 10 | 29 | 25 | 0.124 | 0.089 | 0.087 | 0.200 | 0.141 | 0.110 | 0.228 | 0.199 | 0.056 | 0.123 | undef | 0.103 | 0.213 |
|---|
| cfl1 |   | 8 | 26 | 18 | 16 | 0.216 | 0.134 | 0.083 | -0.084 | 0.142 | 0.251 | -0.466 | 0.229 | 0.174 | 0.188 | 0.208 | -0.025 | 0.215 |
|---|
| prkci |   | 14 | 15 | 31 | 26 | 0.065 | 0.104 | 0.012 | 0.170 | 0.112 | 0.088 | -0.103 | -0.170 | 0.196 | 0.128 | 0.049 | 0.124 | 0.149 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| hras |   | 14 | 15 | 42 | 34 | 0.049 | -0.030 | 0.094 | 0.210 | 0.160 | 0.195 | 0.022 | 0.245 | 0.086 | 0.208 | 0.180 | 0.003 | 0.197 |
|---|
| lgals1 |   | 1 | 116 | 102 | 113 | 0.019 | 0.210 | 0.056 | 0.107 | 0.055 | -0.028 | 0.061 | 0.110 | 0.068 | -0.052 | 0.108 | -0.031 | 0.157 |
|---|
| hsph1 |   | 8 | 26 | 18 | 16 | 0.071 | 0.260 | 0.138 | 0.050 | 0.148 | 0.197 | -0.116 | 0.161 | 0.143 | 0.132 | -0.006 | 0.094 | 0.357 |
|---|
| tesk1 |   | 7 | 30 | 18 | 19 | 0.110 | 0.169 | 0.161 | 0.179 | 0.096 | 0.138 | -0.154 | 0.245 | 0.155 | 0.092 | 0.067 | 0.029 | 0.200 |
|---|
| spry4 |   | 5 | 40 | 18 | 23 | -0.033 | 0.158 | -0.001 | -0.039 | 0.103 | 0.074 | -0.328 | 0.022 | 0.197 | 0.156 | undef | 0.184 | 0.211 |
|---|
| map2k1 |   | 10 | 20 | 54 | 44 | 0.154 | 0.096 | 0.093 | 0.013 | 0.095 | 0.045 | -0.223 | 0.207 | 0.082 | 0.096 | undef | undef | 0.230 |
|---|
GO Enrichment output for subnetwork 3956-4-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.578E-08 | 5.93E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 2.158E-07 | 2.481E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.23E-07 | 3.243E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.429E-06 | 8.217E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.898E-06 | 1.333E-03 |
|---|
| sperm motility | GO:0030317 |  | 5.126E-06 | 1.965E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 9.211E-06 | 3.026E-03 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 1.163E-05 | 3.342E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.247E-05 | 3.186E-03 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 1.501E-05 | 3.453E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 1.501E-05 | 3.139E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| establishment of cell polarity | GO:0030010 |  | 2.031E-10 | 4.961E-07 |
|---|
| polyol catabolic process | GO:0046174 |  | 5.419E-09 | 6.619E-06 |
|---|
| neural plate morphogenesis | GO:0001839 |  | 5.419E-09 | 4.413E-06 |
|---|
| neural plate development | GO:0001840 |  | 1.083E-08 | 6.616E-06 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 4.084E-08 | 1.996E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.543E-07 | 6.282E-05 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.016E-07 | 1.052E-04 |
|---|
| actin filament organization | GO:0007015 |  | 4.075E-07 | 1.244E-04 |
|---|
| neural crest cell migration | GO:0001755 |  | 4.389E-07 | 1.191E-04 |
|---|
| regulation of actin cytoskeleton organization | GO:0032956 |  | 5.069E-07 | 1.238E-04 |
|---|
| regulation of actin filament-based process | GO:0032970 |  | 5.926E-07 | 1.316E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 6.418E-09 | 1.544E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 7.678E-08 | 9.237E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.827E-07 | 1.465E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.57E-07 | 2.148E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.284E-06 | 6.18E-04 |
|---|
| sperm motility | GO:0030317 |  | 1.458E-06 | 5.849E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 3.132E-06 | 1.077E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 4.498E-06 | 1.353E-03 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 4.892E-06 | 1.308E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 6.695E-06 | 1.611E-03 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 7.206E-06 | 1.576E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| establishment of cell polarity | GO:0030010 |  | 2.031E-10 | 4.961E-07 |
|---|
| polyol catabolic process | GO:0046174 |  | 5.419E-09 | 6.619E-06 |
|---|
| neural plate morphogenesis | GO:0001839 |  | 5.419E-09 | 4.413E-06 |
|---|
| neural plate development | GO:0001840 |  | 1.083E-08 | 6.616E-06 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 4.084E-08 | 1.996E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.543E-07 | 6.282E-05 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.016E-07 | 1.052E-04 |
|---|
| actin filament organization | GO:0007015 |  | 4.075E-07 | 1.244E-04 |
|---|
| neural crest cell migration | GO:0001755 |  | 4.389E-07 | 1.191E-04 |
|---|
| regulation of actin cytoskeleton organization | GO:0032956 |  | 5.069E-07 | 1.238E-04 |
|---|
| regulation of actin filament-based process | GO:0032970 |  | 5.926E-07 | 1.316E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 6.418E-09 | 1.544E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 7.678E-08 | 9.237E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.827E-07 | 1.465E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.57E-07 | 2.148E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.284E-06 | 6.18E-04 |
|---|
| sperm motility | GO:0030317 |  | 1.458E-06 | 5.849E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 3.132E-06 | 1.077E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 4.498E-06 | 1.353E-03 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 4.892E-06 | 1.308E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 6.695E-06 | 1.611E-03 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 7.206E-06 | 1.576E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| establishment of cell polarity | GO:0030010 |  | 6.416E-08 | 2.291E-04 |
|---|
| cell cortex | GO:0005938 |  | 3.234E-07 | 5.774E-04 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 1.309E-06 | 1.558E-03 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 2.883E-06 | 2.574E-03 |
|---|
| neural plate development | GO:0001840 |  | 7.176E-06 | 5.125E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.176E-06 | 4.271E-03 |
|---|
| cell cortex part | GO:0044448 |  | 8.508E-06 | 4.34E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.076E-05 | 4.802E-03 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 1.076E-05 | 4.269E-03 |
|---|
| establishment of apical/basal cell polarity | GO:0035089 |  | 1.076E-05 | 3.842E-03 |
|---|
| actin filament organization | GO:0007015 |  | 1.448E-05 | 4.7E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| establishment of cell polarity | GO:0030010 |  | 2.79E-08 | 1.018E-04 |
|---|
| cell cortex | GO:0005938 |  | 1.744E-07 | 3.182E-04 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 5.5E-07 | 6.688E-04 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 1.203E-06 | 1.097E-03 |
|---|
| neural plate morphogenesis | GO:0001839 |  | 5.187E-06 | 3.785E-03 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 5.187E-06 | 3.154E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 5.187E-06 | 2.703E-03 |
|---|
| cell cortex part | GO:0044448 |  | 5.646E-06 | 2.575E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.777E-06 | 3.152E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 7.777E-06 | 2.837E-03 |
|---|
| neural plate development | GO:0001840 |  | 7.777E-06 | 2.579E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 6.418E-09 | 1.544E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 7.678E-08 | 9.237E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.827E-07 | 1.465E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.57E-07 | 2.148E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.284E-06 | 6.18E-04 |
|---|
| sperm motility | GO:0030317 |  | 1.458E-06 | 5.849E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 3.132E-06 | 1.077E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 4.498E-06 | 1.353E-03 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 4.892E-06 | 1.308E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 6.695E-06 | 1.611E-03 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 7.206E-06 | 1.576E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.578E-08 | 5.93E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 2.158E-07 | 2.481E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.23E-07 | 3.243E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.429E-06 | 8.217E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.898E-06 | 1.333E-03 |
|---|
| sperm motility | GO:0030317 |  | 5.126E-06 | 1.965E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 9.211E-06 | 3.026E-03 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 1.163E-05 | 3.342E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.247E-05 | 3.186E-03 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 1.501E-05 | 3.453E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 1.501E-05 | 3.139E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.578E-08 | 5.93E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 2.158E-07 | 2.481E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.23E-07 | 3.243E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.429E-06 | 8.217E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.898E-06 | 1.333E-03 |
|---|
| sperm motility | GO:0030317 |  | 5.126E-06 | 1.965E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 9.211E-06 | 3.026E-03 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 1.163E-05 | 3.342E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.247E-05 | 3.186E-03 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 1.501E-05 | 3.453E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 1.501E-05 | 3.139E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| establishment of cell polarity | GO:0030010 |  | 2.208E-08 | 4.07E-05 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 2.489E-06 | 2.294E-03 |
|---|
| muscle cell differentiation | GO:0042692 |  | 1.293E-05 | 7.941E-03 |
|---|
| regulation of long-term neuronal synaptic plasticity | GO:0048169 |  | 1.897E-05 | 8.741E-03 |
|---|
| establishment or maintenance of epithelial cell apical/basal polarity | GO:0045197 |  | 1.897E-05 | 6.993E-03 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 1.897E-05 | 5.828E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.844E-05 | 7.487E-03 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 2.844E-05 | 6.551E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 2.844E-05 | 5.823E-03 |
|---|
| establishment or maintenance of apical/basal cell polarity | GO:0035088 |  | 2.844E-05 | 5.241E-03 |
|---|
| regulation of synaptic transmission. GABAergic | GO:0032228 |  | 2.844E-05 | 4.764E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.578E-08 | 5.93E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 2.158E-07 | 2.481E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.23E-07 | 3.243E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.429E-06 | 8.217E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.898E-06 | 1.333E-03 |
|---|
| sperm motility | GO:0030317 |  | 5.126E-06 | 1.965E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 9.211E-06 | 3.026E-03 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 1.163E-05 | 3.342E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.247E-05 | 3.186E-03 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 1.501E-05 | 3.453E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 1.501E-05 | 3.139E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.752E-08 | 4.157E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 2.093E-07 | 2.484E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.831E-07 | 3.031E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.179E-06 | 6.996E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.66E-06 | 1.262E-03 |
|---|
| sperm motility | GO:0030317 |  | 4.476E-06 | 1.77E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 7.706E-06 | 2.612E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.023E-05 | 3.035E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 1.325E-05 | 3.495E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.514E-05 | 3.592E-03 |
|---|
| establishment of apical/basal cell polarity | GO:0035089 |  | 1.514E-05 | 3.265E-03 |
|---|



