Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 3008-4-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1817 | 1.656e-02 | 1.327e-02 | 1.294e-01 |
|---|
| IPC-NIBC-129 | 0.2770 | 7.960e-02 | 1.120e-01 | 5.825e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2253 | 4.970e-04 | 1.062e-03 | 4.012e-02 |
|---|
| Loi_GPL570 | 0.2366 | 7.067e-02 | 7.965e-02 | 2.859e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1379 | 0.000e+00 | 6.000e-06 | 0.000e+00 |
|---|
| Parker_GPL1390 | 0.3763 | 1.860e-02 | 2.415e-02 | 2.199e-01 |
|---|
| Parker_GPL887 | 0.1808 | 4.019e-01 | 4.209e-01 | 4.441e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3847 | 2.507e-01 | 1.861e-01 | 8.188e-01 |
|---|
| Schmidt | 0.3113 | 2.929e-02 | 3.472e-02 | 3.007e-01 |
|---|
| Sotiriou | 0.3987 | 2.828e-03 | 2.089e-03 | 8.846e-02 |
|---|
| Van-De-Vijver | 0.2227 | 6.940e-04 | 9.980e-04 | 4.584e-02 |
|---|
| Zhang | 0.2474 | 1.865e-01 | 1.331e-01 | 3.664e-01 |
|---|
| Zhou | 0.4408 | 5.301e-02 | 4.081e-02 | 3.296e-01 |
|---|
Expression data for subnetwork 3008-4-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
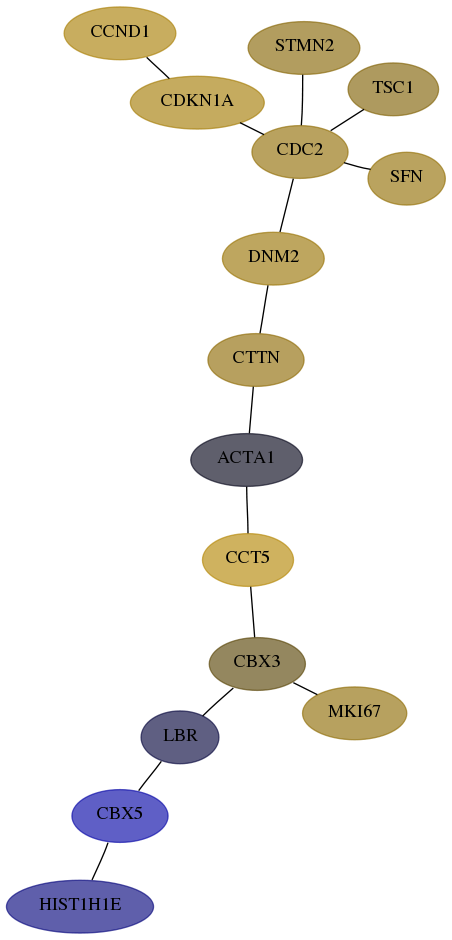
Score for each gene in subnetwork 3008-4-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| cbx3 |   | 1 | 116 | 64 | 74 | 0.018 | 0.130 | 0.050 | 0.062 | 0.108 | 0.185 | 0.071 | 0.092 | 0.205 | 0.128 | -0.040 | 0.086 | 0.285 |
|---|
| stmn2 |   | 29 | 9 | 34 | 27 | 0.057 | 0.106 | 0.005 | 0.052 | 0.110 | 0.071 | -0.099 | 0.006 | 0.034 | 0.103 | 0.008 | 0.113 | -0.039 |
|---|
| cttn |   | 10 | 20 | 1 | 6 | 0.067 | 0.065 | 0.123 | 0.238 | 0.126 | 0.263 | -0.172 | 0.216 | 0.069 | 0.115 | undef | 0.025 | 0.279 |
|---|
| lbr |   | 1 | 116 | 64 | 74 | -0.009 | 0.246 | -0.020 | -0.267 | 0.019 | 0.243 | 0.117 | 0.042 | 0.164 | -0.081 | -0.009 | 0.089 | 0.320 |
|---|
| cdkn1a |   | 44 | 7 | 1 | 4 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | undef | -0.201 | 0.069 |
|---|
| mki67 |   | 1 | 116 | 64 | 74 | 0.069 | 0.259 | 0.138 | 0.190 | 0.141 | 0.042 | 0.203 | 0.260 | 0.238 | 0.224 | 0.111 | 0.224 | 0.210 |
|---|
| cct5 |   | 2 | 78 | 64 | 63 | 0.165 | 0.258 | 0.182 | 0.155 | 0.209 | 0.212 | 0.257 | 0.353 | 0.143 | 0.204 | undef | 0.039 | 0.289 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| hist1h1e |   | 1 | 116 | 64 | 74 | -0.044 | -0.059 | 0.093 | -0.096 | 0.078 | 0.067 | 0.234 | -0.063 | 0.279 | 0.074 | 0.108 | 0.100 | -0.152 |
|---|
| cbx5 |   | 1 | 116 | 64 | 74 | -0.120 | 0.114 | 0.058 | 0.097 | 0.113 | 0.060 | 0.020 | -0.070 | 0.233 | 0.173 | 0.083 | 0.104 | 0.200 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| dnm2 |   | 3 | 62 | 64 | 61 | 0.089 | 0.127 | 0.114 | -0.049 | 0.065 | 0.251 | 0.291 | 0.177 | -0.039 | -0.048 | 0.078 | -0.044 | 0.262 |
|---|
| sfn |   | 61 | 3 | 1 | 2 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.158 | 0.079 | 0.181 |
|---|
| acta1 |   | 9 | 22 | 1 | 7 | -0.003 | 0.098 | -0.008 | -0.030 | 0.119 | 0.174 | 0.376 | 0.024 | 0.182 | 0.214 | 0.100 | 0.048 | 0.201 |
|---|
GO Enrichment output for subnetwork 3008-4-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.766E-07 | 1.786E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.378E-06 | 3.885E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.381E-06 | 4.892E-03 |
|---|
| interphase | GO:0051325 |  | 6.994E-06 | 4.021E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.958E-05 | 9.008E-03 |
|---|
| skeletal myofibril assembly | GO:0014866 |  | 1.958E-05 | 7.506E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.958E-05 | 6.434E-03 |
|---|
| chromatin remodeling | GO:0006338 |  | 2.282E-05 | 6.561E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.465E-05 | 6.299E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.07E-05 | 7.061E-03 |
|---|
| response to UV | GO:0009411 |  | 3.765E-05 | 7.873E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.066E-07 | 5.047E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.504E-06 | 1.837E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.841E-06 | 1.499E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.839E-06 | 4.788E-03 |
|---|
| skeletal myofibril assembly | GO:0014866 |  | 7.839E-06 | 3.83E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 9.166E-06 | 3.732E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.8E-06 | 3.42E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 1.175E-05 | 3.589E-03 |
|---|
| chromatin remodeling | GO:0006338 |  | 1.506E-05 | 4.088E-03 |
|---|
| response to UV | GO:0009411 |  | 1.684E-05 | 4.115E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.191E-05 | 4.866E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.045E-07 | 7.326E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.021E-06 | 2.431E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.39E-06 | 1.917E-03 |
|---|
| interphase | GO:0051325 |  | 2.707E-06 | 1.628E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 9.574E-06 | 4.607E-03 |
|---|
| skeletal myofibril assembly | GO:0014866 |  | 9.574E-06 | 3.839E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 9.574E-06 | 3.291E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.23E-05 | 3.7E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.315E-05 | 3.516E-03 |
|---|
| chromatin remodeling | GO:0006338 |  | 1.798E-05 | 4.327E-03 |
|---|
| response to UV | GO:0009411 |  | 2.02E-05 | 4.419E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.066E-07 | 5.047E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.504E-06 | 1.837E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.841E-06 | 1.499E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.839E-06 | 4.788E-03 |
|---|
| skeletal myofibril assembly | GO:0014866 |  | 7.839E-06 | 3.83E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 9.166E-06 | 3.732E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.8E-06 | 3.42E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 1.175E-05 | 3.589E-03 |
|---|
| chromatin remodeling | GO:0006338 |  | 1.506E-05 | 4.088E-03 |
|---|
| response to UV | GO:0009411 |  | 1.684E-05 | 4.115E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.191E-05 | 4.866E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.045E-07 | 7.326E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.021E-06 | 2.431E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.39E-06 | 1.917E-03 |
|---|
| interphase | GO:0051325 |  | 2.707E-06 | 1.628E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 9.574E-06 | 4.607E-03 |
|---|
| skeletal myofibril assembly | GO:0014866 |  | 9.574E-06 | 3.839E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 9.574E-06 | 3.291E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.23E-05 | 3.7E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.315E-05 | 3.516E-03 |
|---|
| chromatin remodeling | GO:0006338 |  | 1.798E-05 | 4.327E-03 |
|---|
| response to UV | GO:0009411 |  | 2.02E-05 | 4.419E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| general transcriptional repressor activity | GO:0016565 |  | 6.822E-09 | 2.436E-05 |
|---|
| centromeric heterochromatin | GO:0005721 |  | 1.364E-08 | 2.435E-05 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 3.814E-08 | 4.54E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.122E-07 | 1.001E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.145E-07 | 8.181E-05 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 2.471E-07 | 1.47E-04 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 3.136E-07 | 1.6E-04 |
|---|
| nuclear heterochromatin | GO:0005720 |  | 4.608E-07 | 2.057E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.608E-07 | 1.828E-04 |
|---|
| kinase regulator activity | GO:0019207 |  | 6.633E-07 | 2.368E-04 |
|---|
| nuclear inner membrane | GO:0005637 |  | 7.712E-07 | 2.503E-04 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| chromocenter | GO:0010369 |  | 6.213E-09 | 2.267E-05 |
|---|
| general transcriptional repressor activity | GO:0016565 |  | 1.242E-08 | 2.265E-05 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 3.474E-08 | 4.224E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.133E-07 | 1.033E-04 |
|---|
| nuclear heterochromatin | GO:0005720 |  | 1.362E-07 | 9.934E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 2.25E-07 | 1.368E-04 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 3.24E-07 | 1.688E-04 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 3.458E-07 | 1.577E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.458E-07 | 1.402E-04 |
|---|
| kinase regulator activity | GO:0019207 |  | 5.735E-07 | 2.092E-04 |
|---|
| nuclear inner membrane | GO:0005637 |  | 7.024E-07 | 2.329E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.045E-07 | 7.326E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.021E-06 | 2.431E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.39E-06 | 1.917E-03 |
|---|
| interphase | GO:0051325 |  | 2.707E-06 | 1.628E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 9.574E-06 | 4.607E-03 |
|---|
| skeletal myofibril assembly | GO:0014866 |  | 9.574E-06 | 3.839E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 9.574E-06 | 3.291E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.23E-05 | 3.7E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.315E-05 | 3.516E-03 |
|---|
| chromatin remodeling | GO:0006338 |  | 1.798E-05 | 4.327E-03 |
|---|
| response to UV | GO:0009411 |  | 2.02E-05 | 4.419E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.766E-07 | 1.786E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.378E-06 | 3.885E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.381E-06 | 4.892E-03 |
|---|
| interphase | GO:0051325 |  | 6.994E-06 | 4.021E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.958E-05 | 9.008E-03 |
|---|
| skeletal myofibril assembly | GO:0014866 |  | 1.958E-05 | 7.506E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.958E-05 | 6.434E-03 |
|---|
| chromatin remodeling | GO:0006338 |  | 2.282E-05 | 6.561E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.465E-05 | 6.299E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.07E-05 | 7.061E-03 |
|---|
| response to UV | GO:0009411 |  | 3.765E-05 | 7.873E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.766E-07 | 1.786E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.378E-06 | 3.885E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.381E-06 | 4.892E-03 |
|---|
| interphase | GO:0051325 |  | 6.994E-06 | 4.021E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.958E-05 | 9.008E-03 |
|---|
| skeletal myofibril assembly | GO:0014866 |  | 1.958E-05 | 7.506E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.958E-05 | 6.434E-03 |
|---|
| chromatin remodeling | GO:0006338 |  | 2.282E-05 | 6.561E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.465E-05 | 6.299E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.07E-05 | 7.061E-03 |
|---|
| response to UV | GO:0009411 |  | 3.765E-05 | 7.873E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| chromatin assembly or disassembly | GO:0006333 |  | 2.734E-06 | 5.04E-03 |
|---|
| nucleosome positioning | GO:0016584 |  | 2.318E-05 | 0.02136177 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 3.474E-05 | 0.02134313 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 3.474E-05 | 0.01600735 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 3.474E-05 | 0.01280588 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 3.474E-05 | 0.01067157 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 4.86E-05 | 0.01279471 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 4.86E-05 | 0.01119537 |
|---|
| myofibril assembly | GO:0030239 |  | 4.86E-05 | 9.951E-03 |
|---|
| regulation of stress fiber formation | GO:0051492 |  | 6.474E-05 | 0.01193131 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 6.474E-05 | 0.01084665 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.766E-07 | 1.786E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.378E-06 | 3.885E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.381E-06 | 4.892E-03 |
|---|
| interphase | GO:0051325 |  | 6.994E-06 | 4.021E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.958E-05 | 9.008E-03 |
|---|
| skeletal myofibril assembly | GO:0014866 |  | 1.958E-05 | 7.506E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.958E-05 | 6.434E-03 |
|---|
| chromatin remodeling | GO:0006338 |  | 2.282E-05 | 6.561E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.465E-05 | 6.299E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.07E-05 | 7.061E-03 |
|---|
| response to UV | GO:0009411 |  | 3.765E-05 | 7.873E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.016E-06 | 2.411E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.253E-06 | 1.487E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.642E-06 | 2.881E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 3.642E-06 | 2.161E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.463E-06 | 3.542E-03 |
|---|
| interphase | GO:0051325 |  | 8.169E-06 | 3.231E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.047E-05 | 6.941E-03 |
|---|
| skeletal myofibril assembly | GO:0014866 |  | 2.047E-05 | 6.073E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 3.068E-05 | 8.09E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.08E-05 | 7.309E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.546E-05 | 7.65E-03 |
|---|



