Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 2885-3-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1943 | 1.242e-02 | 9.579e-03 | 1.035e-01 |
|---|
| IPC-NIBC-129 | 0.2830 | 1.738e-02 | 2.603e-02 | 2.082e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2393 | 8.231e-03 | 1.393e-02 | 2.445e-01 |
|---|
| Loi_GPL570 | 0.2280 | 7.974e-02 | 8.956e-02 | 3.130e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1548 | 0.000e+00 | 4.000e-06 | 0.000e+00 |
|---|
| Parker_GPL1390 | 0.2488 | 4.164e-02 | 5.217e-02 | 3.559e-01 |
|---|
| Parker_GPL887 | 0.0153 | 3.723e-01 | 3.912e-01 | 4.077e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3789 | 9.670e-01 | 9.585e-01 | 1.000e+00 |
|---|
| Schmidt | 0.2557 | 1.752e-02 | 2.009e-02 | 2.137e-01 |
|---|
| Sotiriou | 0.3447 | 7.042e-03 | 5.332e-03 | 1.537e-01 |
|---|
| Van-De-Vijver | 0.3332 | 1.171e-03 | 1.686e-03 | 6.522e-02 |
|---|
| Zhang | 0.2787 | 1.451e-01 | 9.706e-02 | 2.992e-01 |
|---|
| Zhou | 0.4115 | 5.589e-02 | 4.374e-02 | 3.435e-01 |
|---|
Expression data for subnetwork 2885-3-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
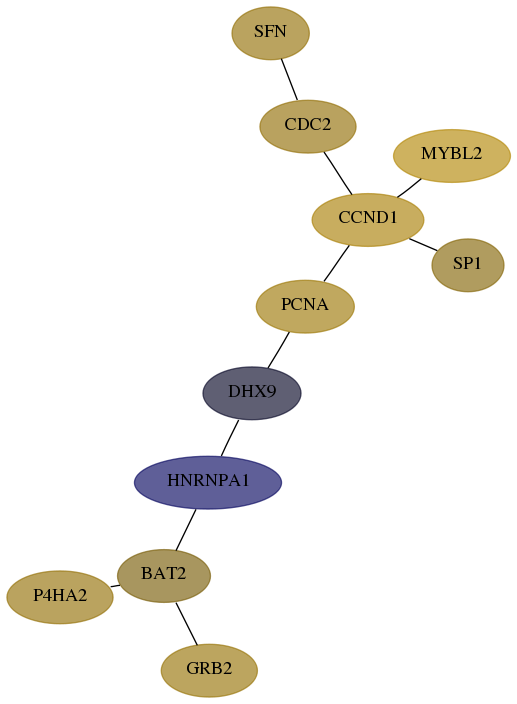
Score for each gene in subnetwork 2885-3-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| pcna |   | 15 | 11 | 110 | 85 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.119 | 0.104 | 0.256 | 0.151 | 0.059 | 0.201 | 0.123 | 0.233 |
|---|
| mybl2 |   | 9 | 22 | 56 | 48 | 0.162 | 0.237 | 0.166 | 0.079 | 0.124 | 0.184 | 0.333 | 0.275 | 0.202 | 0.182 | 0.289 | 0.074 | 0.245 |
|---|
| bat2 |   | 3 | 62 | 74 | 68 | 0.039 | 0.182 | -0.001 | -0.013 | 0.062 | undef | -0.335 | 0.109 | 0.186 | 0.099 | undef | 0.059 | 0.146 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| dhx9 |   | 1 | 116 | 129 | 136 | -0.004 | 0.165 | 0.099 | -0.094 | 0.103 | undef | -0.029 | 0.127 | 0.155 | 0.135 | 0.094 | 0.007 | 0.234 |
|---|
| sp1 |   | 8 | 26 | 71 | 57 | 0.054 | 0.186 | 0.147 | 0.242 | 0.125 | 0.033 | -0.370 | 0.202 | 0.284 | 0.227 | 0.304 | 0.091 | -0.068 |
|---|
| hnrnpa1 |   | 1 | 116 | 129 | 136 | -0.021 | 0.209 | 0.088 | -0.020 | 0.087 | 0.131 | undef | 0.082 | 0.036 | 0.089 | undef | 0.083 | 0.195 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| p4ha2 |   | 3 | 62 | 74 | 68 | 0.077 | 0.313 | 0.070 | 0.159 | 0.149 | 0.216 | 0.056 | 0.180 | 0.055 | 0.105 | 0.211 | 0.043 | 0.237 |
|---|
| sfn |   | 61 | 3 | 1 | 2 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.158 | 0.079 | 0.181 |
|---|
| grb2 |   | 9 | 22 | 1 | 7 | 0.084 | -0.013 | 0.086 | -0.031 | 0.162 | -0.038 | 0.003 | 0.065 | 0.158 | 0.127 | 0.149 | 0.194 | -0.005 |
|---|
GO Enrichment output for subnetwork 2885-3-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.266E-05 | 0.02911443 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.851E-05 | 0.02128154 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.096E-05 | 0.0160718 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 2.793E-05 | 0.01606199 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 6.568E-05 | 0.03021449 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 9.045E-05 | 0.0346738 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 9.045E-05 | 0.0297204 |
|---|
| positive regulation of cell size | GO:0045793 |  | 1.043E-04 | 0.02998787 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.043E-04 | 0.02665588 |
|---|
| skin development | GO:0043588 |  | 1.043E-04 | 0.0239903 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.191E-04 | 0.02490978 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.593E-06 | 6.334E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 3.005E-06 | 3.67E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.372E-06 | 3.56E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 4.506E-06 | 2.752E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 8.405E-06 | 4.107E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 1.649E-05 | 6.715E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 2.337E-05 | 8.158E-03 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 2.726E-05 | 8.325E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.726E-05 | 7.4E-03 |
|---|
| positive regulation of cell size | GO:0045793 |  | 3.592E-05 | 8.776E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.592E-05 | 7.979E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 4.185E-06 | 0.01006818 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.249E-06 | 5.112E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 6.274E-06 | 5.032E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.162E-06 | 4.308E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.17E-05 | 5.631E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 1.504E-05 | 6.031E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 2.754E-05 | 9.466E-03 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 3.794E-05 | 0.01141156 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 3.794E-05 | 0.0101436 |
|---|
| positive regulation of cell size | GO:0045793 |  | 4.376E-05 | 0.01052955 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 5E-05 | 0.01093543 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.593E-06 | 6.334E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 3.005E-06 | 3.67E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.372E-06 | 3.56E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 4.506E-06 | 2.752E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 8.405E-06 | 4.107E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 1.649E-05 | 6.715E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 2.337E-05 | 8.158E-03 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 2.726E-05 | 8.325E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.726E-05 | 7.4E-03 |
|---|
| positive regulation of cell size | GO:0045793 |  | 3.592E-05 | 8.776E-03 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 3.592E-05 | 7.979E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 4.185E-06 | 0.01006818 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.249E-06 | 5.112E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 6.274E-06 | 5.032E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.162E-06 | 4.308E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.17E-05 | 5.631E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 1.504E-05 | 6.031E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 2.754E-05 | 9.466E-03 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 3.794E-05 | 0.01141156 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 3.794E-05 | 0.0101436 |
|---|
| positive regulation of cell size | GO:0045793 |  | 4.376E-05 | 0.01052955 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 5E-05 | 0.01093543 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 8.812E-09 | 3.147E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.045E-06 | 5.437E-03 |
|---|
| embryonic camera-type eye development | GO:0031076 |  | 3.314E-06 | 3.944E-03 |
|---|
| MutLalpha complex binding | GO:0032405 |  | 3.314E-06 | 2.958E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.969E-06 | 3.549E-03 |
|---|
| insulin receptor substrate binding | GO:0043560 |  | 4.969E-06 | 2.957E-03 |
|---|
| enucleate erythrocyte differentiation | GO:0043353 |  | 4.969E-06 | 2.535E-03 |
|---|
| superoxide-generating NADPH oxidase activity | GO:0016175 |  | 4.969E-06 | 2.218E-03 |
|---|
| DNA replication factor C complex | GO:0005663 |  | 4.969E-06 | 1.972E-03 |
|---|
| definitive hemopoiesis | GO:0060216 |  | 4.969E-06 | 1.774E-03 |
|---|
| procollagen-proline dioxygenase activity | GO:0019798 |  | 4.969E-06 | 1.613E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.462E-08 | 5.334E-05 |
|---|
| enucleate erythrocyte differentiation | GO:0043353 |  | 4.447E-06 | 8.111E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 4.447E-06 | 5.407E-03 |
|---|
| epidermal growth factor receptor binding | GO:0005154 |  | 4.447E-06 | 4.055E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.344E-06 | 3.899E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.667E-06 | 4.054E-03 |
|---|
| embryonic camera-type eye development | GO:0031076 |  | 6.667E-06 | 3.475E-03 |
|---|
| MutLalpha complex binding | GO:0032405 |  | 6.667E-06 | 3.04E-03 |
|---|
| DNA replication factor C complex | GO:0005663 |  | 6.667E-06 | 2.702E-03 |
|---|
| insulin receptor substrate binding | GO:0043560 |  | 9.33E-06 | 3.404E-03 |
|---|
| procollagen-proline dioxygenase activity | GO:0019798 |  | 9.33E-06 | 3.094E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 4.185E-06 | 0.01006818 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.249E-06 | 5.112E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 6.274E-06 | 5.032E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.162E-06 | 4.308E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.17E-05 | 5.631E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 1.504E-05 | 6.031E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 2.754E-05 | 9.466E-03 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 3.794E-05 | 0.01141156 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 3.794E-05 | 0.0101436 |
|---|
| positive regulation of cell size | GO:0045793 |  | 4.376E-05 | 0.01052955 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 5E-05 | 0.01093543 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.266E-05 | 0.02911443 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.851E-05 | 0.02128154 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.096E-05 | 0.0160718 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 2.793E-05 | 0.01606199 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 6.568E-05 | 0.03021449 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 9.045E-05 | 0.0346738 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 9.045E-05 | 0.0297204 |
|---|
| positive regulation of cell size | GO:0045793 |  | 1.043E-04 | 0.02998787 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.043E-04 | 0.02665588 |
|---|
| skin development | GO:0043588 |  | 1.043E-04 | 0.0239903 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.191E-04 | 0.02490978 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.266E-05 | 0.02911443 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.851E-05 | 0.02128154 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.096E-05 | 0.0160718 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 2.793E-05 | 0.01606199 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 6.568E-05 | 0.03021449 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 9.045E-05 | 0.0346738 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 9.045E-05 | 0.0297204 |
|---|
| positive regulation of cell size | GO:0045793 |  | 1.043E-04 | 0.02998787 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.043E-04 | 0.02665588 |
|---|
| skin development | GO:0043588 |  | 1.043E-04 | 0.0239903 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.191E-04 | 0.02490978 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell growth | GO:0030307 |  | 1.771E-05 | 0.03263475 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 2.478E-05 | 0.02283104 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 3.302E-05 | 0.02028245 |
|---|
| positive regulation of cell size | GO:0045793 |  | 4.242E-05 | 0.0195467 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 4.242E-05 | 0.01563736 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 4.242E-05 | 0.01303113 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 5.3E-05 | 0.01395381 |
|---|
| skin development | GO:0043588 |  | 6.474E-05 | 0.01491414 |
|---|
| negative regulation of peptidase activity | GO:0010466 |  | 7.764E-05 | 0.01589916 |
|---|
| regulation of chromosome organization | GO:0033044 |  | 7.764E-05 | 0.01430924 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 7.764E-05 | 0.0130084 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.266E-05 | 0.02911443 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.851E-05 | 0.02128154 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.096E-05 | 0.0160718 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 2.793E-05 | 0.01606199 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 6.568E-05 | 0.03021449 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 9.045E-05 | 0.0346738 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 9.045E-05 | 0.0297204 |
|---|
| positive regulation of cell size | GO:0045793 |  | 1.043E-04 | 0.02998787 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 1.043E-04 | 0.02665588 |
|---|
| skin development | GO:0043588 |  | 1.043E-04 | 0.0239903 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 1.191E-04 | 0.02490978 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| RNA export from nucleus | GO:0006405 |  | 1.338E-07 | 3.174E-04 |
|---|
| nuclear export | GO:0051168 |  | 5.154E-07 | 6.116E-04 |
|---|
| nuclear import | GO:0051170 |  | 3.251E-06 | 2.572E-03 |
|---|
| regulation of integrin biosynthetic process | GO:0045113 |  | 1.277E-05 | 7.574E-03 |
|---|
| definitive hemopoiesis | GO:0060216 |  | 1.277E-05 | 6.059E-03 |
|---|
| embryonic camera-type eye development | GO:0031076 |  | 1.914E-05 | 7.568E-03 |
|---|
| response to pH | GO:0009268 |  | 1.914E-05 | 6.487E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.677E-05 | 7.941E-03 |
|---|
| enucleate erythrocyte differentiation | GO:0043353 |  | 2.677E-05 | 7.059E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.048E-05 | 7.232E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 3.567E-05 | 7.695E-03 |
|---|



