Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 2178-2-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1999 | 1.567e-02 | 1.247e-02 | 1.240e-01 |
|---|
| IPC-NIBC-129 | 0.3039 | 4.456e-02 | 6.446e-02 | 4.135e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2280 | 4.324e-03 | 7.731e-03 | 1.677e-01 |
|---|
| Loi_GPL570 | 0.2099 | 1.025e-01 | 1.143e-01 | 3.757e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1538 | 0.000e+00 | 1.000e-06 | 0.000e+00 |
|---|
| Parker_GPL1390 | 0.2783 | 5.478e-02 | 6.776e-02 | 4.140e-01 |
|---|
| Parker_GPL887 | 0.0849 | 3.598e-01 | 3.786e-01 | 3.923e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3907 | 6.157e-01 | 5.497e-01 | 9.875e-01 |
|---|
| Schmidt | 0.2366 | 1.592e-02 | 1.813e-02 | 1.999e-01 |
|---|
| Sotiriou | 0.3114 | 1.229e-02 | 9.461e-03 | 2.118e-01 |
|---|
| Van-De-Vijver | 0.2653 | 1.153e-03 | 1.659e-03 | 6.455e-02 |
|---|
| Zhang | 0.2845 | 1.474e-01 | 9.903e-02 | 3.031e-01 |
|---|
| Zhou | 0.4124 | 5.015e-02 | 3.794e-02 | 3.155e-01 |
|---|
Expression data for subnetwork 2178-2-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
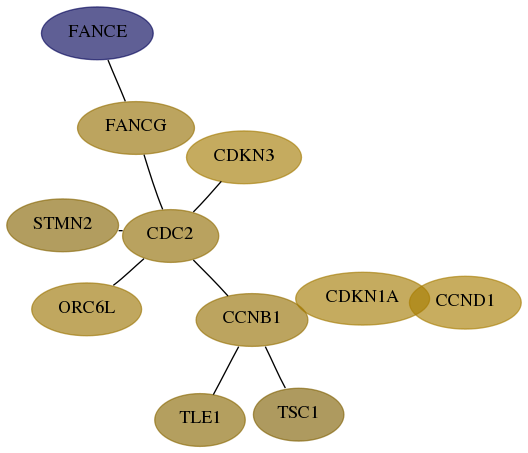
Score for each gene in subnetwork 2178-2-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| cdkn3 |   | 53 | 5 | 29 | 22 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.166 | -0.009 | 0.323 | 0.186 | 0.198 | 0.201 | 0.271 | 0.372 |
|---|
| fance |   | 1 | 116 | 137 | 141 | -0.019 | 0.105 | 0.076 | 0.206 | 0.079 | 0.241 | 0.126 | 0.135 | 0.209 | 0.052 | undef | 0.104 | 0.023 |
|---|
| stmn2 |   | 29 | 9 | 34 | 27 | 0.057 | 0.106 | 0.005 | 0.052 | 0.110 | 0.071 | -0.099 | 0.006 | 0.034 | 0.103 | 0.008 | 0.113 | -0.039 |
|---|
| cdkn1a |   | 44 | 7 | 1 | 4 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | undef | -0.201 | 0.069 |
|---|
| tle1 |   | 47 | 6 | 1 | 3 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | 0.170 | 0.068 | 0.025 |
|---|
| fancg |   | 1 | 116 | 137 | 141 | 0.081 | 0.214 | 0.140 | 0.118 | 0.100 | 0.152 | 0.239 | 0.215 | 0.173 | 0.118 | 0.075 | 0.070 | 0.344 |
|---|
| ccnb1 |   | 42 | 8 | 56 | 39 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | undef | 0.167 | 0.398 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| orc6l |   | 15 | 11 | 34 | 28 | 0.094 | 0.270 | 0.084 | 0.114 | 0.106 | 0.240 | 0.256 | 0.234 | 0.192 | 0.105 | 0.258 | 0.175 | 0.160 |
|---|
GO Enrichment output for subnetwork 2178-2-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.513E-08 | 3.479E-05 |
|---|
| interphase | GO:0051325 |  | 1.7E-08 | 1.955E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.376E-08 | 5.655E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.487E-07 | 8.548E-05 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.244E-07 | 1.952E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 9.247E-07 | 3.545E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.005E-06 | 3.302E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.005E-06 | 2.889E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.336E-06 | 3.415E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.885E-06 | 2.044E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 8.885E-06 | 1.858E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.824E-09 | 6.898E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.445E-08 | 1.766E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.484E-08 | 2.837E-05 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.013E-07 | 1.229E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 2.54E-07 | 1.241E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.054E-07 | 1.65E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 4.581E-07 | 1.599E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 6.43E-07 | 1.964E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.321E-06 | 9.015E-04 |
|---|
| retrograde protein transport. ER to cytosol | GO:0030970 |  | 3.321E-06 | 8.113E-04 |
|---|
| response to UV | GO:0009411 |  | 4.571E-06 | 1.015E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.192E-09 | 1.009E-05 |
|---|
| interphase | GO:0051325 |  | 4.908E-09 | 5.904E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.266E-08 | 1.817E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.46E-08 | 3.284E-05 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.41E-07 | 1.16E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 3.974E-07 | 1.594E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 5.706E-07 | 1.961E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 5.706E-07 | 1.716E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 8.117E-07 | 2.17E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 4.185E-06 | 1.007E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 4.185E-06 | 9.153E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.824E-09 | 6.898E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.445E-08 | 1.766E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.484E-08 | 2.837E-05 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.013E-07 | 1.229E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 2.54E-07 | 1.241E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.054E-07 | 1.65E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 4.581E-07 | 1.599E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 6.43E-07 | 1.964E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.321E-06 | 9.015E-04 |
|---|
| retrograde protein transport. ER to cytosol | GO:0030970 |  | 3.321E-06 | 8.113E-04 |
|---|
| response to UV | GO:0009411 |  | 4.571E-06 | 1.015E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.192E-09 | 1.009E-05 |
|---|
| interphase | GO:0051325 |  | 4.908E-09 | 5.904E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.266E-08 | 1.817E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.46E-08 | 3.284E-05 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.41E-07 | 1.16E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 3.974E-07 | 1.594E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 5.706E-07 | 1.961E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 5.706E-07 | 1.716E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 8.117E-07 | 2.17E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 4.185E-06 | 1.007E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 4.185E-06 | 9.153E-04 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.163E-09 | 4.154E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.259E-08 | 2.247E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.722E-08 | 2.05E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.427E-08 | 2.167E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 3.704E-08 | 2.646E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.523E-07 | 9.066E-05 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.169E-07 | 1.107E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 5.806E-07 | 2.592E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 7.31E-07 | 2.901E-04 |
|---|
| response to UV | GO:0009411 |  | 4.081E-06 | 1.457E-03 |
|---|
| retrograde protein transport. ER to cytosol | GO:0030970 |  | 4.142E-06 | 1.344E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.035E-09 | 3.775E-06 |
|---|
| interphase | GO:0051325 |  | 1.172E-09 | 2.138E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.125E-08 | 1.368E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.178E-08 | 1.075E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.38E-08 | 1.736E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 7.296E-08 | 4.436E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.122E-07 | 5.846E-05 |
|---|
| response to UV | GO:0009411 |  | 2.813E-06 | 1.283E-03 |
|---|
| origin recognition complex | GO:0000808 |  | 3.763E-06 | 1.525E-03 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 3.763E-06 | 1.373E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 3.763E-06 | 1.248E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.192E-09 | 1.009E-05 |
|---|
| interphase | GO:0051325 |  | 4.908E-09 | 5.904E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.266E-08 | 1.817E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.46E-08 | 3.284E-05 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 2.41E-07 | 1.16E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 3.974E-07 | 1.594E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 5.706E-07 | 1.961E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 5.706E-07 | 1.716E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 8.117E-07 | 2.17E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 4.185E-06 | 1.007E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 4.185E-06 | 9.153E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.513E-08 | 3.479E-05 |
|---|
| interphase | GO:0051325 |  | 1.7E-08 | 1.955E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.376E-08 | 5.655E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.487E-07 | 8.548E-05 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.244E-07 | 1.952E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 9.247E-07 | 3.545E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.005E-06 | 3.302E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.005E-06 | 2.889E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.336E-06 | 3.415E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.885E-06 | 2.044E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 8.885E-06 | 1.858E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.513E-08 | 3.479E-05 |
|---|
| interphase | GO:0051325 |  | 1.7E-08 | 1.955E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.376E-08 | 5.655E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.487E-07 | 8.548E-05 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.244E-07 | 1.952E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 9.247E-07 | 3.545E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.005E-06 | 3.302E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.005E-06 | 2.889E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.336E-06 | 3.415E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.885E-06 | 2.044E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 8.885E-06 | 1.858E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 9.493E-06 | 0.01749647 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 9.493E-06 | 8.748E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 9.493E-06 | 5.832E-03 |
|---|
| proteasomal protein catabolic process | GO:0010498 |  | 1.302E-05 | 5.998E-03 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 1.329E-05 | 4.897E-03 |
|---|
| regulation of stress fiber formation | GO:0051492 |  | 1.771E-05 | 5.439E-03 |
|---|
| negative regulation of signal transduction | GO:0009968 |  | 2.06E-05 | 5.425E-03 |
|---|
| positive regulation of hydrolase activity | GO:0051345 |  | 2.149E-05 | 4.952E-03 |
|---|
| regulation of dephosphorylation | GO:0035303 |  | 2.276E-05 | 4.66E-03 |
|---|
| regulation of actin filament bundle formation | GO:0032231 |  | 2.276E-05 | 4.194E-03 |
|---|
| negative regulation of cell communication | GO:0010648 |  | 2.53E-05 | 4.239E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.513E-08 | 3.479E-05 |
|---|
| interphase | GO:0051325 |  | 1.7E-08 | 1.955E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.376E-08 | 5.655E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.487E-07 | 8.548E-05 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.244E-07 | 1.952E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 9.247E-07 | 3.545E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.005E-06 | 3.302E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.005E-06 | 2.889E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.336E-06 | 3.415E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.885E-06 | 2.044E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 8.885E-06 | 1.858E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.168E-08 | 2.772E-05 |
|---|
| interphase | GO:0051325 |  | 1.311E-08 | 1.555E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.232E-08 | 4.93E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.338E-07 | 7.935E-05 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 5.037E-07 | 2.39E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 8.195E-07 | 3.241E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 9.083E-07 | 3.079E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.245E-06 | 3.693E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.598E-06 | 4.212E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.724E-06 | 1.833E-03 |
|---|
| response to UV | GO:0009411 |  | 9.845E-06 | 2.124E-03 |
|---|



