Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 2139-2-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1387 | 1.831e-02 | 1.486e-02 | 1.397e-01 |
|---|
| IPC-NIBC-129 | 0.2960 | 5.313e-02 | 7.625e-02 | 4.621e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2204 | 5.933e-03 | 1.033e-02 | 2.025e-01 |
|---|
| Loi_GPL570 | 0.2120 | 9.950e-02 | 1.111e-01 | 3.678e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1718 | 0.000e+00 | 2.000e-06 | 0.000e+00 |
|---|
| Parker_GPL1390 | 0.2638 | 5.599e-02 | 6.919e-02 | 4.188e-01 |
|---|
| Parker_GPL887 | 0.0518 | 5.186e-01 | 5.373e-01 | 5.850e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3775 | 7.920e-01 | 7.481e-01 | 9.985e-01 |
|---|
| Schmidt | 0.2351 | 1.964e-02 | 2.270e-02 | 2.311e-01 |
|---|
| Sotiriou | 0.2730 | 2.318e-02 | 1.820e-02 | 2.992e-01 |
|---|
| Van-De-Vijver | 0.2525 | 1.150e-03 | 1.655e-03 | 6.444e-02 |
|---|
| Zhang | 0.2718 | 1.123e-01 | 7.036e-02 | 2.421e-01 |
|---|
| Zhou | 0.4125 | 5.660e-02 | 4.447e-02 | 3.469e-01 |
|---|
Expression data for subnetwork 2139-2-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
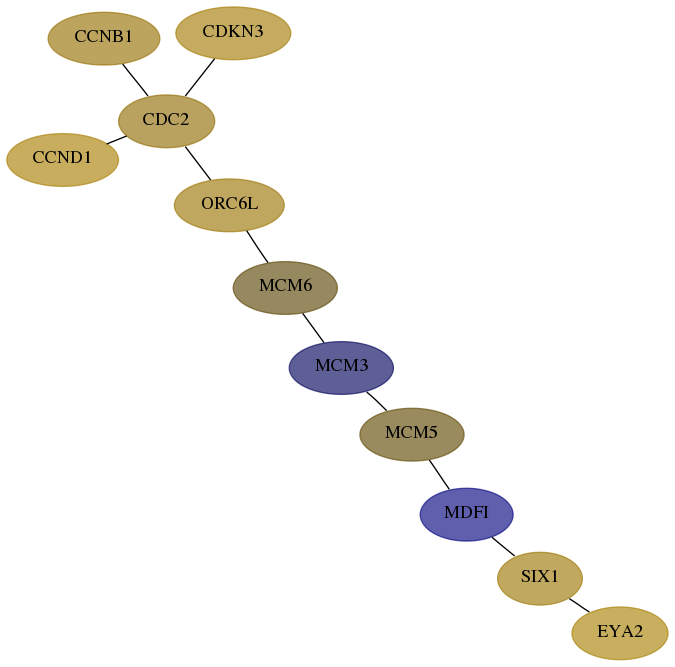
Score for each gene in subnetwork 2139-2-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| cdkn3 |   | 53 | 5 | 29 | 22 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.166 | -0.009 | 0.323 | 0.186 | 0.198 | 0.201 | 0.271 | 0.372 |
|---|
| mcm3 |   | 1 | 116 | 253 | 255 | -0.020 | 0.173 | 0.113 | 0.084 | 0.110 | 0.125 | 0.151 | 0.255 | 0.171 | 0.156 | 0.174 | -0.003 | 0.335 |
|---|
| mcm5 |   | 1 | 116 | 253 | 255 | 0.023 | 0.121 | 0.170 | 0.015 | 0.075 | 0.209 | 0.117 | 0.228 | 0.198 | 0.011 | 0.168 | 0.053 | 0.185 |
|---|
| eya2 |   | 1 | 116 | 253 | 255 | 0.131 | 0.170 | 0.034 | -0.002 | 0.090 | 0.258 | -0.408 | -0.084 | 0.062 | 0.206 | undef | -0.034 | 0.109 |
|---|
| ccnb1 |   | 42 | 8 | 56 | 39 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | undef | 0.167 | 0.398 |
|---|
| six1 |   | 13 | 17 | 123 | 96 | 0.097 | 0.114 | 0.095 | 0.149 | 0.151 | -0.015 | -0.006 | 0.123 | 0.113 | 0.184 | 0.082 | 0.191 | 0.079 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| mcm6 |   | 1 | 116 | 253 | 255 | 0.021 | 0.271 | 0.148 | 0.061 | 0.121 | 0.090 | -0.055 | 0.320 | 0.258 | 0.144 | 0.134 | 0.084 | 0.283 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| orc6l |   | 15 | 11 | 34 | 28 | 0.094 | 0.270 | 0.084 | 0.114 | 0.106 | 0.240 | 0.256 | 0.234 | 0.192 | 0.105 | 0.258 | 0.175 | 0.160 |
|---|
| mdfi |   | 7 | 30 | 42 | 35 | -0.050 | 0.215 | 0.036 | -0.005 | 0.054 | 0.263 | 0.176 | 0.130 | 0.296 | -0.041 | 0.074 | 0.076 | 0.056 |
|---|
GO Enrichment output for subnetwork 2139-2-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA replication initiation | GO:0006270 |  | 6.347E-10 | 1.46E-06 |
|---|
| DNA-dependent DNA replication | GO:0006261 |  | 4.246E-08 | 4.883E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.149E-07 | 5.481E-04 |
|---|
| interphase | GO:0051325 |  | 7.842E-07 | 4.509E-04 |
|---|
| cell fate specification | GO:0001708 |  | 2.264E-06 | 1.042E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.178E-06 | 2.368E-03 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 1.029E-05 | 3.38E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.038E-05 | 2.985E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 3.679E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.785E-05 | 4.106E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.875E-05 | 3.92E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA replication initiation | GO:0006270 |  | 1.057E-10 | 2.581E-07 |
|---|
| DNA-dependent DNA replication | GO:0006261 |  | 1.553E-08 | 1.897E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.04E-07 | 1.661E-04 |
|---|
| cell fate specification | GO:0001708 |  | 7.668E-07 | 4.683E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.937E-06 | 9.465E-04 |
|---|
| muscle cell migration | GO:0014812 |  | 2.705E-06 | 1.101E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.72E-06 | 1.298E-03 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 4.055E-06 | 1.238E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 6.348E-06 | 1.723E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 6.635E-06 | 1.621E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 7.545E-06 | 1.676E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA replication initiation | GO:0006270 |  | 1.618E-10 | 3.892E-07 |
|---|
| DNA-dependent DNA replication | GO:0006261 |  | 1.86E-08 | 2.238E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.748E-07 | 2.204E-04 |
|---|
| interphase | GO:0051325 |  | 3.114E-07 | 1.873E-04 |
|---|
| cell fate specification | GO:0001708 |  | 9.671E-07 | 4.654E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.678E-06 | 1.074E-03 |
|---|
| muscle cell migration | GO:0014812 |  | 3.37E-06 | 1.158E-03 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 5.053E-06 | 1.52E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.14E-06 | 1.374E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 8.768E-06 | 2.109E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 9.163E-06 | 2.004E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA replication initiation | GO:0006270 |  | 1.057E-10 | 2.581E-07 |
|---|
| DNA-dependent DNA replication | GO:0006261 |  | 1.553E-08 | 1.897E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.04E-07 | 1.661E-04 |
|---|
| cell fate specification | GO:0001708 |  | 7.668E-07 | 4.683E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.937E-06 | 9.465E-04 |
|---|
| muscle cell migration | GO:0014812 |  | 2.705E-06 | 1.101E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.72E-06 | 1.298E-03 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 4.055E-06 | 1.238E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 6.348E-06 | 1.723E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 6.635E-06 | 1.621E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 7.545E-06 | 1.676E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA replication initiation | GO:0006270 |  | 1.618E-10 | 3.892E-07 |
|---|
| DNA-dependent DNA replication | GO:0006261 |  | 1.86E-08 | 2.238E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.748E-07 | 2.204E-04 |
|---|
| interphase | GO:0051325 |  | 3.114E-07 | 1.873E-04 |
|---|
| cell fate specification | GO:0001708 |  | 9.671E-07 | 4.654E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.678E-06 | 1.074E-03 |
|---|
| muscle cell migration | GO:0014812 |  | 3.37E-06 | 1.158E-03 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 5.053E-06 | 1.52E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.14E-06 | 1.374E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 8.768E-06 | 2.109E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 9.163E-06 | 2.004E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA replication initiation | GO:0006270 |  | 1.067E-10 | 3.812E-07 |
|---|
| DNA-dependent DNA replication | GO:0006261 |  | 7.101E-08 | 1.268E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.824E-07 | 2.171E-04 |
|---|
| cell fate specification | GO:0001708 |  | 9.053E-07 | 8.082E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.366E-06 | 2.404E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.339E-06 | 2.583E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.21E-06 | 3.168E-03 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 6.21E-06 | 2.772E-03 |
|---|
| alpha DNA polymerase:primase complex | GO:0005658 |  | 6.21E-06 | 2.464E-03 |
|---|
| nuclear origin of replication recognition complex | GO:0005664 |  | 6.21E-06 | 2.218E-03 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.158E-05 | 3.76E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA replication initiation | GO:0006270 |  | 1.316E-10 | 4.8E-07 |
|---|
| DNA-dependent DNA replication | GO:0006261 |  | 1.422E-08 | 2.593E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.568E-07 | 1.907E-04 |
|---|
| interphase | GO:0051325 |  | 1.731E-07 | 1.578E-04 |
|---|
| cell fate specification | GO:0001708 |  | 1.815E-06 | 1.325E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.448E-06 | 1.488E-03 |
|---|
| origin recognition complex | GO:0000808 |  | 3.763E-06 | 1.961E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 3.763E-06 | 1.716E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.119E-06 | 1.67E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 5.642E-06 | 2.058E-03 |
|---|
| alpha DNA polymerase:primase complex | GO:0005658 |  | 5.642E-06 | 1.871E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA replication initiation | GO:0006270 |  | 1.618E-10 | 3.892E-07 |
|---|
| DNA-dependent DNA replication | GO:0006261 |  | 1.86E-08 | 2.238E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.748E-07 | 2.204E-04 |
|---|
| interphase | GO:0051325 |  | 3.114E-07 | 1.873E-04 |
|---|
| cell fate specification | GO:0001708 |  | 9.671E-07 | 4.654E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.678E-06 | 1.074E-03 |
|---|
| muscle cell migration | GO:0014812 |  | 3.37E-06 | 1.158E-03 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 5.053E-06 | 1.52E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.14E-06 | 1.374E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 8.768E-06 | 2.109E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 9.163E-06 | 2.004E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA replication initiation | GO:0006270 |  | 6.347E-10 | 1.46E-06 |
|---|
| DNA-dependent DNA replication | GO:0006261 |  | 4.246E-08 | 4.883E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.149E-07 | 5.481E-04 |
|---|
| interphase | GO:0051325 |  | 7.842E-07 | 4.509E-04 |
|---|
| cell fate specification | GO:0001708 |  | 2.264E-06 | 1.042E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.178E-06 | 2.368E-03 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 1.029E-05 | 3.38E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.038E-05 | 2.985E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 3.679E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.785E-05 | 4.106E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.875E-05 | 3.92E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA replication initiation | GO:0006270 |  | 6.347E-10 | 1.46E-06 |
|---|
| DNA-dependent DNA replication | GO:0006261 |  | 4.246E-08 | 4.883E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.149E-07 | 5.481E-04 |
|---|
| interphase | GO:0051325 |  | 7.842E-07 | 4.509E-04 |
|---|
| cell fate specification | GO:0001708 |  | 2.264E-06 | 1.042E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.178E-06 | 2.368E-03 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 1.029E-05 | 3.38E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.038E-05 | 2.985E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 3.679E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.785E-05 | 4.106E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.875E-05 | 3.92E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA replication initiation | GO:0006270 |  | 4.72E-11 | 8.698E-08 |
|---|
| DNA replication | GO:0006260 |  | 7.456E-09 | 6.87E-06 |
|---|
| DNA-dependent DNA replication | GO:0006261 |  | 8.835E-09 | 5.427E-06 |
|---|
| muscle cell migration | GO:0014812 |  | 8.861E-06 | 4.083E-03 |
|---|
| cytoplasmic sequestering of transcription factor | GO:0042994 |  | 1.329E-05 | 4.897E-03 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 1.329E-05 | 4.081E-03 |
|---|
| thymus development | GO:0048538 |  | 1.329E-05 | 3.498E-03 |
|---|
| axis specification | GO:0009798 |  | 1.859E-05 | 4.283E-03 |
|---|
| induction of an organ | GO:0001759 |  | 1.859E-05 | 3.807E-03 |
|---|
| thyroid gland development | GO:0030878 |  | 2.478E-05 | 4.566E-03 |
|---|
| negative regulation of transcription factor import into nucleus | GO:0042992 |  | 2.478E-05 | 4.151E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA replication initiation | GO:0006270 |  | 6.347E-10 | 1.46E-06 |
|---|
| DNA-dependent DNA replication | GO:0006261 |  | 4.246E-08 | 4.883E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.149E-07 | 5.481E-04 |
|---|
| interphase | GO:0051325 |  | 7.842E-07 | 4.509E-04 |
|---|
| cell fate specification | GO:0001708 |  | 2.264E-06 | 1.042E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.178E-06 | 2.368E-03 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 1.029E-05 | 3.38E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.038E-05 | 2.985E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 3.679E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.785E-05 | 4.106E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.875E-05 | 3.92E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA replication initiation | GO:0006270 |  | 3.792E-10 | 8.999E-07 |
|---|
| DNA-dependent DNA replication | GO:0006261 |  | 3.997E-08 | 4.743E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.494E-07 | 3.555E-04 |
|---|
| interphase | GO:0051325 |  | 4.925E-07 | 2.922E-04 |
|---|
| cell fate specification | GO:0001708 |  | 2.048E-06 | 9.72E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.512E-06 | 1.785E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.949E-06 | 2.695E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.113E-05 | 3.3E-03 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 1.113E-05 | 2.934E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.544E-05 | 3.663E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.843E-05 | 3.976E-03 |
|---|



