Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 2079-2-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2444 | 5.388e-03 | 3.707e-03 | 5.278e-02 |
|---|
| IPC-NIBC-129 | 0.2326 | 3.308e-02 | 4.844e-02 | 3.380e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2798 | 7.761e-03 | 1.320e-02 | 2.365e-01 |
|---|
| Loi_GPL570 | 0.2621 | 4.929e-02 | 5.610e-02 | 2.160e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1543 | 1.000e-05 | 8.700e-05 | 6.000e-05 |
|---|
| Parker_GPL1390 | 0.2515 | 9.962e-02 | 1.196e-01 | 5.598e-01 |
|---|
| Parker_GPL887 | 0.0233 | 2.729e-01 | 2.904e-01 | 2.850e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.4068 | 9.355e-01 | 9.196e-01 | 1.000e+00 |
|---|
| Schmidt | 0.1944 | 3.352e-01 | 4.174e-01 | 9.139e-01 |
|---|
| Sotiriou | 0.3480 | 6.660e-03 | 5.035e-03 | 1.488e-01 |
|---|
| Van-De-Vijver | 0.2870 | 1.408e-03 | 2.026e-03 | 7.366e-02 |
|---|
| Zhang | 0.0920 | 1.463e-01 | 9.807e-02 | 3.012e-01 |
|---|
| Zhou | 0.4012 | 4.319e-02 | 3.114e-02 | 2.796e-01 |
|---|
Expression data for subnetwork 2079-2-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
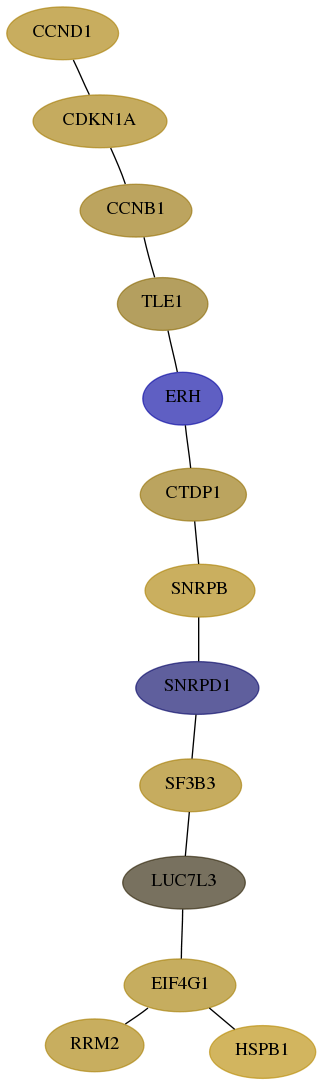
Score for each gene in subnetwork 2079-2-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| ctdp1 |   | 1 | 116 | 251 | 253 | 0.080 | 0.063 | 0.111 | -0.001 | 0.089 | 0.158 | 0.003 | 0.121 | -0.065 | 0.044 | 0.137 | 0.144 | 0.010 |
|---|
| rrm2 |   | 5 | 40 | 163 | 143 | 0.131 | 0.207 | 0.227 | 0.187 | 0.161 | 0.178 | 0.137 | 0.303 | 0.190 | 0.219 | undef | 0.129 | 0.302 |
|---|
| cdkn1a |   | 44 | 7 | 1 | 4 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | undef | -0.201 | 0.069 |
|---|
| tle1 |   | 47 | 6 | 1 | 3 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | 0.170 | 0.068 | 0.025 |
|---|
| snrpd1 |   | 2 | 78 | 196 | 190 | -0.026 | 0.257 | 0.128 | 0.254 | 0.106 | 0.174 | 0.255 | 0.155 | 0.190 | 0.111 | 0.083 | 0.011 | 0.205 |
|---|
| luc7l3 |   | 2 | 78 | 196 | 190 | 0.006 | 0.111 | -0.046 | -0.016 | 0.121 | 0.071 | -0.318 | -0.084 | 0.159 | 0.098 | -0.004 | 0.067 | 0.065 |
|---|
| ccnb1 |   | 42 | 8 | 56 | 39 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | undef | 0.167 | 0.398 |
|---|
| eif4g1 |   | 2 | 78 | 196 | 190 | 0.123 | 0.262 | 0.043 | 0.013 | 0.098 | 0.189 | 0.016 | 0.194 | 0.076 | 0.147 | 0.208 | -0.028 | 0.268 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| hspb1 |   | 5 | 40 | 42 | 36 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | -0.026 | -0.065 | 0.289 | -0.021 | 0.201 | 0.183 | -0.077 | 0.290 |
|---|
| snrpb |   | 3 | 62 | 1 | 14 | 0.141 | 0.077 | 0.167 | 0.250 | 0.139 | 0.089 | 0.155 | 0.281 | 0.120 | 0.161 | 0.167 | 0.010 | 0.153 |
|---|
| sf3b3 |   | 3 | 62 | 196 | 184 | 0.119 | 0.241 | 0.143 | -0.019 | 0.082 | 0.222 | 0.007 | 0.271 | 0.168 | 0.060 | 0.123 | -0.077 | 0.372 |
|---|
| erh |   | 1 | 116 | 251 | 253 | -0.108 | 0.077 | 0.128 | 0.178 | 0.073 | -0.067 | -0.337 | 0.046 | 0.085 | 0.034 | -0.036 | 0.230 | 0.232 |
|---|
GO Enrichment output for subnetwork 2079-2-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.451E-08 | 7.937E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.034E-08 | 6.939E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.267E-06 | 1.738E-03 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.267E-06 | 1.304E-03 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 2.623E-06 | 1.206E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.732E-06 | 1.43E-03 |
|---|
| interphase | GO:0051325 |  | 4.091E-06 | 1.344E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.066E-05 | 5.94E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 2.372E-05 | 6.061E-03 |
|---|
| response to UV | GO:0009411 |  | 2.535E-05 | 5.831E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.163E-05 | 6.613E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.573E-07 | 1.606E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 6.997E-07 | 8.547E-04 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 9.952E-07 | 8.104E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.573E-06 | 2.793E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 7.445E-06 | 3.638E-03 |
|---|
| response to UV | GO:0009411 |  | 7.871E-06 | 3.205E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.77E-06 | 3.061E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 9.952E-06 | 3.039E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.326E-05 | 3.6E-03 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.705E-05 | 4.164E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.13E-05 | 4.73E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.994E-08 | 4.798E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.781E-08 | 5.752E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 8.7E-07 | 6.978E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.297E-06 | 7.802E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.317E-06 | 6.336E-04 |
|---|
| interphase | GO:0051325 |  | 1.491E-06 | 5.981E-04 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 1.648E-06 | 5.663E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.459E-06 | 2.544E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.3E-05 | 3.476E-03 |
|---|
| response to UV | GO:0009411 |  | 1.3E-05 | 3.128E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.62E-05 | 3.544E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.573E-07 | 1.606E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 6.997E-07 | 8.547E-04 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 9.952E-07 | 8.104E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.573E-06 | 2.793E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 7.445E-06 | 3.638E-03 |
|---|
| response to UV | GO:0009411 |  | 7.871E-06 | 3.205E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.77E-06 | 3.061E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 9.952E-06 | 3.039E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.326E-05 | 3.6E-03 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.705E-05 | 4.164E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.13E-05 | 4.73E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.994E-08 | 4.798E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.781E-08 | 5.752E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 8.7E-07 | 6.978E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.297E-06 | 7.802E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.317E-06 | 6.336E-04 |
|---|
| interphase | GO:0051325 |  | 1.491E-06 | 5.981E-04 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 1.648E-06 | 5.663E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.459E-06 | 2.544E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.3E-05 | 3.476E-03 |
|---|
| response to UV | GO:0009411 |  | 1.3E-05 | 3.128E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.62E-05 | 3.544E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| small nuclear ribonucleoprotein complex | GO:0030532 |  | 4.561E-09 | 1.629E-05 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 2.997E-08 | 5.352E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 8.818E-08 | 1.05E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.623E-07 | 3.234E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.132E-07 | 4.38E-04 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 1.935E-06 | 1.152E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 5.61E-06 | 2.862E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.969E-06 | 3.557E-03 |
|---|
| response to UV | GO:0009411 |  | 9.657E-06 | 3.832E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.027E-05 | 3.667E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.076E-05 | 3.492E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| small nuclear ribonucleoprotein complex | GO:0030532 |  | 2.062E-09 | 7.521E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.861E-08 | 3.394E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.206E-07 | 1.467E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.854E-07 | 1.691E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.15E-07 | 2.298E-04 |
|---|
| interphase | GO:0051325 |  | 3.476E-07 | 2.114E-04 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 9.637E-07 | 5.022E-04 |
|---|
| regulation of translational initiation | GO:0006446 |  | 2.766E-06 | 1.261E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.037E-06 | 1.636E-03 |
|---|
| response to UV | GO:0009411 |  | 4.638E-06 | 1.692E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.789E-06 | 2.251E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.994E-08 | 4.798E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.781E-08 | 5.752E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 8.7E-07 | 6.978E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.297E-06 | 7.802E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.317E-06 | 6.336E-04 |
|---|
| interphase | GO:0051325 |  | 1.491E-06 | 5.981E-04 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 1.648E-06 | 5.663E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.459E-06 | 2.544E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.3E-05 | 3.476E-03 |
|---|
| response to UV | GO:0009411 |  | 1.3E-05 | 3.128E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.62E-05 | 3.544E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.451E-08 | 7.937E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.034E-08 | 6.939E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.267E-06 | 1.738E-03 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.267E-06 | 1.304E-03 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 2.623E-06 | 1.206E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.732E-06 | 1.43E-03 |
|---|
| interphase | GO:0051325 |  | 4.091E-06 | 1.344E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.066E-05 | 5.94E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 2.372E-05 | 6.061E-03 |
|---|
| response to UV | GO:0009411 |  | 2.535E-05 | 5.831E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.163E-05 | 6.613E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.451E-08 | 7.937E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.034E-08 | 6.939E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.267E-06 | 1.738E-03 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.267E-06 | 1.304E-03 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 2.623E-06 | 1.206E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.732E-06 | 1.43E-03 |
|---|
| interphase | GO:0051325 |  | 4.091E-06 | 1.344E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.066E-05 | 5.94E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 2.372E-05 | 6.061E-03 |
|---|
| response to UV | GO:0009411 |  | 2.535E-05 | 5.831E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.163E-05 | 6.613E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 5.739E-07 | 1.058E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 4.569E-06 | 4.211E-03 |
|---|
| regulation of translation | GO:0006417 |  | 5.216E-05 | 0.03204601 |
|---|
| negative regulation of Wnt receptor signaling pathway | GO:0030178 |  | 9.17E-05 | 0.04225272 |
|---|
| pyrimidine nucleoside metabolic process | GO:0006213 |  | 9.17E-05 | 0.03380217 |
|---|
| posttranscriptional regulation of gene expression | GO:0010608 |  | 1.49E-04 | 0.04577903 |
|---|
| response to heat | GO:0009408 |  | 2.003E-04 | 0.05274745 |
|---|
| spliceosome assembly | GO:0000245 |  | 2.457E-04 | 0.05661443 |
|---|
| regulation of Wnt receptor signaling pathway | GO:0030111 |  | 2.702E-04 | 0.05532411 |
|---|
| response to temperature stimulus | GO:0009266 |  | 5.064E-04 | 0.09332788 |
|---|
| nucleoside metabolic process | GO:0009116 |  | 5.41E-04 | 0.09064201 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.451E-08 | 7.937E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.034E-08 | 6.939E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.267E-06 | 1.738E-03 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.267E-06 | 1.304E-03 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 2.623E-06 | 1.206E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.732E-06 | 1.43E-03 |
|---|
| interphase | GO:0051325 |  | 4.091E-06 | 1.344E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.066E-05 | 5.94E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 2.372E-05 | 6.061E-03 |
|---|
| response to UV | GO:0009411 |  | 2.535E-05 | 5.831E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.163E-05 | 6.613E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 2.343E-08 | 5.56E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.714E-08 | 5.594E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 7.537E-08 | 5.962E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.773E-06 | 1.052E-03 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.051E-06 | 9.734E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.839E-06 | 1.123E-03 |
|---|
| interphase | GO:0051325 |  | 3.109E-06 | 1.054E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.735E-05 | 5.146E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.985E-05 | 5.235E-03 |
|---|
| response to UV | GO:0009411 |  | 2.119E-05 | 5.029E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.677E-05 | 5.775E-03 |
|---|



