Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1912-2-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1767 | 1.971e-02 | 1.614e-02 | 1.477e-01 |
|---|
| IPC-NIBC-129 | 0.2805 | 5.157e-02 | 7.411e-02 | 4.537e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2168 | 4.370e-04 | 9.430e-04 | 3.660e-02 |
|---|
| Loi_GPL570 | 0.3779 | 9.127e-03 | 1.082e-02 | 5.032e-02 |
|---|
| Loi_GPL96-GPL97 | 0.1262 | 0.000e+00 | 5.000e-06 | 0.000e+00 |
|---|
| Parker_GPL1390 | 0.3821 | 1.392e-02 | 1.831e-02 | 1.829e-01 |
|---|
| Parker_GPL887 | 0.4154 | 4.142e-01 | 4.333e-01 | 4.592e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3462 | 1.879e-02 | 8.589e-03 | 1.687e-01 |
|---|
| Schmidt | 0.3310 | 3.190e-01 | 3.983e-01 | 9.042e-01 |
|---|
| Sotiriou | 0.2723 | 2.347e-02 | 1.843e-02 | 3.012e-01 |
|---|
| Van-De-Vijver | 0.2547 | 2.846e-03 | 4.084e-03 | 1.157e-01 |
|---|
| Zhang | 0.0954 | 2.214e-01 | 1.649e-01 | 4.191e-01 |
|---|
| Zhou | 0.3617 | 7.541e-02 | 6.462e-02 | 4.290e-01 |
|---|
Expression data for subnetwork 1912-2-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
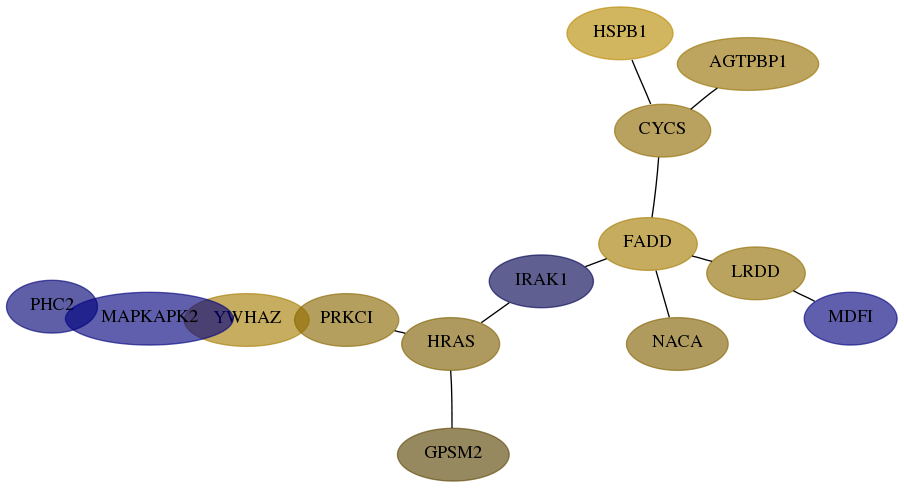
Score for each gene in subnetwork 1912-2-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| agtpbp1 |   | 1 | 116 | 42 | 58 | 0.086 | 0.219 | 0.009 | 0.181 | 0.071 | 0.078 | 0.296 | 0.026 | 0.110 | 0.151 | 0.042 | 0.064 | 0.226 |
|---|
| irak1 |   | 5 | 40 | 42 | 36 | -0.016 | 0.242 | 0.007 | 0.133 | 0.108 | 0.303 | 0.257 | 0.133 | 0.177 | 0.084 | 0.199 | -0.026 | 0.081 |
|---|
| cycs |   | 1 | 116 | 42 | 58 | 0.073 | 0.206 | 0.080 | 0.094 | 0.117 | -0.056 | 0.202 | 0.272 | 0.157 | 0.077 | undef | 0.135 | 0.234 |
|---|
| ywhaz |   | 17 | 10 | 29 | 25 | 0.124 | 0.089 | 0.087 | 0.200 | 0.141 | 0.110 | 0.228 | 0.199 | 0.056 | 0.123 | undef | 0.103 | 0.213 |
|---|
| prkci |   | 14 | 15 | 31 | 26 | 0.065 | 0.104 | 0.012 | 0.170 | 0.112 | 0.088 | -0.103 | -0.170 | 0.196 | 0.128 | 0.049 | 0.124 | 0.149 |
|---|
| fadd |   | 4 | 50 | 42 | 40 | 0.121 | -0.009 | 0.155 | 0.307 | 0.166 | 0.119 | 0.084 | 0.300 | 0.070 | 0.116 | 0.051 | 0.032 | 0.243 |
|---|
| hspb1 |   | 5 | 40 | 42 | 36 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | -0.026 | -0.065 | 0.289 | -0.021 | 0.201 | 0.183 | -0.077 | 0.290 |
|---|
| gpsm2 |   | 4 | 50 | 42 | 40 | 0.021 | 0.286 | 0.155 | 0.144 | 0.108 | 0.242 | 0.307 | 0.134 | 0.234 | 0.077 | 0.112 | 0.098 | 0.207 |
|---|
| hras |   | 14 | 15 | 42 | 34 | 0.049 | -0.030 | 0.094 | 0.210 | 0.160 | 0.195 | 0.022 | 0.245 | 0.086 | 0.208 | 0.180 | 0.003 | 0.197 |
|---|
| phc2 |   | 1 | 116 | 42 | 58 | -0.039 | 0.115 | 0.031 | -0.011 | 0.054 | 0.156 | 0.184 | -0.036 | -0.083 | -0.013 | 0.095 | -0.102 | 0.193 |
|---|
| mapkapk2 |   | 3 | 62 | 42 | 45 | -0.050 | 0.102 | 0.156 | -0.116 | 0.090 | 0.168 | 0.303 | 0.074 | 0.171 | 0.117 | 0.092 | -0.124 | 0.312 |
|---|
| mdfi |   | 7 | 30 | 42 | 35 | -0.050 | 0.215 | 0.036 | -0.005 | 0.054 | 0.263 | 0.176 | 0.130 | 0.296 | -0.041 | 0.074 | 0.076 | 0.056 |
|---|
| lrdd |   | 3 | 62 | 42 | 45 | 0.075 | 0.117 | -0.015 | 0.179 | 0.068 | 0.006 | 0.302 | 0.104 | 0.152 | 0.007 | undef | 0.108 | 0.085 |
|---|
| naca |   | 3 | 62 | 42 | 45 | 0.052 | 0.141 | 0.119 | 0.132 | 0.102 | -0.017 | -0.128 | 0.081 | 0.076 | 0.100 | -0.184 | 0.147 | 0.138 |
|---|
GO Enrichment output for subnetwork 1912-2-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.578E-08 | 5.93E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.23E-07 | 4.865E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.429E-06 | 1.096E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.898E-06 | 1.666E-03 |
|---|
| sperm motility | GO:0030317 |  | 5.126E-06 | 2.358E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 9.211E-06 | 3.531E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.247E-05 | 4.096E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 1.501E-05 | 4.316E-03 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 2.108E-05 | 5.388E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 2.935E-05 | 6.75E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.935E-05 | 6.136E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 5.999E-09 | 1.466E-05 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 3.354E-08 | 4.097E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.708E-07 | 1.391E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.338E-07 | 2.039E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 4.051E-07 | 1.979E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 7.904E-07 | 3.218E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 9.147E-07 | 3.192E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 1.201E-06 | 3.667E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.541E-06 | 4.182E-04 |
|---|
| sperm motility | GO:0030317 |  | 1.939E-06 | 4.737E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 2.929E-06 | 6.504E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 8.016E-09 | 1.929E-05 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 4.48E-08 | 5.39E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.281E-07 | 1.829E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 4.457E-07 | 2.681E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 5.409E-07 | 2.603E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.055E-06 | 4.231E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.221E-06 | 4.197E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 1.603E-06 | 4.82E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.603E-06 | 4.284E-04 |
|---|
| sperm motility | GO:0030317 |  | 1.82E-06 | 4.379E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 3.907E-06 | 8.546E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 5.999E-09 | 1.466E-05 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 3.354E-08 | 4.097E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.708E-07 | 1.391E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.338E-07 | 2.039E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 4.051E-07 | 1.979E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 7.904E-07 | 3.218E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 9.147E-07 | 3.192E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 1.201E-06 | 3.667E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.541E-06 | 4.182E-04 |
|---|
| sperm motility | GO:0030317 |  | 1.939E-06 | 4.737E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 2.929E-06 | 6.504E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 8.016E-09 | 1.929E-05 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 4.48E-08 | 5.39E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.281E-07 | 1.829E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 4.457E-07 | 2.681E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 5.409E-07 | 2.603E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.055E-06 | 4.231E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.221E-06 | 4.197E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 1.603E-06 | 4.82E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.603E-06 | 4.284E-04 |
|---|
| sperm motility | GO:0030317 |  | 1.82E-06 | 4.379E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 3.907E-06 | 8.546E-04 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| death receptor binding | GO:0005123 |  | 3.814E-08 | 1.362E-04 |
|---|
| tumor necrosis factor receptor superfamily binding | GO:0032813 |  | 2.459E-06 | 4.391E-03 |
|---|
| lipopolysaccharide-mediated signaling pathway | GO:0031663 |  | 1.255E-05 | 0.01493743 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 1.255E-05 | 0.01120307 |
|---|
| establishment of apical/basal cell polarity | GO:0035089 |  | 1.255E-05 | 8.962E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.756E-05 | 0.01045052 |
|---|
| eye photoreceptor cell differentiation | GO:0001754 |  | 1.756E-05 | 8.958E-03 |
|---|
| regulation of synaptic transmission. GABAergic | GO:0032228 |  | 1.756E-05 | 7.838E-03 |
|---|
| positive regulation of small GTPase mediated signal transduction | GO:0051057 |  | 2.34E-05 | 9.284E-03 |
|---|
| regulation of long-term neuronal synaptic plasticity | GO:0048169 |  | 2.34E-05 | 8.356E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 3.007E-05 | 9.761E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| death receptor binding | GO:0005123 |  | 2.861E-08 | 1.044E-04 |
|---|
| tumor necrosis factor receptor superfamily binding | GO:0032813 |  | 1.847E-06 | 3.368E-03 |
|---|
| NF-kappaB-inducing kinase activity | GO:0004704 |  | 6.839E-06 | 8.316E-03 |
|---|
| regulation of synaptic transmission. GABAergic | GO:0032228 |  | 1.025E-05 | 9.351E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.435E-05 | 0.01046752 |
|---|
| dorsal/ventral axis specification | GO:0009950 |  | 1.435E-05 | 8.723E-03 |
|---|
| cerebellar Purkinje cell layer development | GO:0021680 |  | 1.435E-05 | 7.477E-03 |
|---|
| regulation of long-term neuronal synaptic plasticity | GO:0048169 |  | 1.435E-05 | 6.542E-03 |
|---|
| establishment or maintenance of epithelial cell apical/basal polarity | GO:0045197 |  | 1.435E-05 | 5.815E-03 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 1.435E-05 | 5.234E-03 |
|---|
| cerebellar cortex formation | GO:0021697 |  | 1.912E-05 | 6.341E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 8.016E-09 | 1.929E-05 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 4.48E-08 | 5.39E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.281E-07 | 1.829E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 4.457E-07 | 2.681E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 5.409E-07 | 2.603E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.055E-06 | 4.231E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.221E-06 | 4.197E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 1.603E-06 | 4.82E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.603E-06 | 4.284E-04 |
|---|
| sperm motility | GO:0030317 |  | 1.82E-06 | 4.379E-04 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 3.907E-06 | 8.546E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.578E-08 | 5.93E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.23E-07 | 4.865E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.429E-06 | 1.096E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.898E-06 | 1.666E-03 |
|---|
| sperm motility | GO:0030317 |  | 5.126E-06 | 2.358E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 9.211E-06 | 3.531E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.247E-05 | 4.096E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 1.501E-05 | 4.316E-03 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 2.108E-05 | 5.388E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 2.935E-05 | 6.75E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.935E-05 | 6.136E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.578E-08 | 5.93E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.23E-07 | 4.865E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.429E-06 | 1.096E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.898E-06 | 1.666E-03 |
|---|
| sperm motility | GO:0030317 |  | 5.126E-06 | 2.358E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 9.211E-06 | 3.531E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.247E-05 | 4.096E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 1.501E-05 | 4.316E-03 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 2.108E-05 | 5.388E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 2.935E-05 | 6.75E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.935E-05 | 6.136E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of long-term neuronal synaptic plasticity | GO:0048169 |  | 2.318E-05 | 0.04272354 |
|---|
| establishment or maintenance of epithelial cell apical/basal polarity | GO:0045197 |  | 2.318E-05 | 0.02136177 |
|---|
| activation of NF-kappaB-inducing kinase activity | GO:0007250 |  | 2.318E-05 | 0.01424118 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 2.318E-05 | 0.01068088 |
|---|
| establishment of cell polarity | GO:0030010 |  | 2.318E-05 | 8.545E-03 |
|---|
| cytoplasmic sequestering of transcription factor | GO:0042994 |  | 3.474E-05 | 0.01067157 |
|---|
| establishment or maintenance of apical/basal cell polarity | GO:0035088 |  | 3.474E-05 | 9.147E-03 |
|---|
| regulation of synaptic transmission. GABAergic | GO:0032228 |  | 3.474E-05 | 8.004E-03 |
|---|
| axis specification | GO:0009798 |  | 4.86E-05 | 9.951E-03 |
|---|
| visual behavior | GO:0007632 |  | 4.86E-05 | 8.956E-03 |
|---|
| regulation of neuronal synaptic plasticity | GO:0048168 |  | 6.474E-05 | 0.01084665 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.578E-08 | 5.93E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.23E-07 | 4.865E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.429E-06 | 1.096E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.898E-06 | 1.666E-03 |
|---|
| sperm motility | GO:0030317 |  | 5.126E-06 | 2.358E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 9.211E-06 | 3.531E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.247E-05 | 4.096E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 1.501E-05 | 4.316E-03 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 2.108E-05 | 5.388E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 2.935E-05 | 6.75E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.935E-05 | 6.136E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.542E-08 | 3.658E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.373E-07 | 4.002E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.038E-06 | 8.213E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 2.342E-06 | 1.39E-03 |
|---|
| sperm motility | GO:0030317 |  | 3.942E-06 | 1.871E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 6.788E-06 | 2.685E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 9.015E-06 | 3.056E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 1.168E-05 | 3.464E-03 |
|---|
| establishment of apical/basal cell polarity | GO:0035089 |  | 1.393E-05 | 3.672E-03 |
|---|
| alcohol biosynthetic process | GO:0046165 |  | 1.481E-05 | 3.515E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.087E-05 | 4.503E-03 |
|---|



