Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1854-2-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2077 | 1.152e-02 | 8.791e-03 | 9.751e-02 |
|---|
| IPC-NIBC-129 | 0.2477 | 2.981e-02 | 4.382e-02 | 3.139e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2430 | 2.877e-02 | 4.358e-02 | 4.645e-01 |
|---|
| Loi_GPL570 | 0.2199 | 8.920e-02 | 9.988e-02 | 3.399e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1814 | 3.000e-06 | 3.500e-05 | 1.000e-05 |
|---|
| Parker_GPL1390 | 0.1907 | 7.640e-02 | 9.298e-02 | 4.921e-01 |
|---|
| Parker_GPL887 | 0.0563 | 3.429e-01 | 3.615e-01 | 3.714e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.4019 | 7.674e-01 | 7.196e-01 | 9.979e-01 |
|---|
| Schmidt | 0.2132 | 2.447e-02 | 2.869e-02 | 2.676e-01 |
|---|
| Sotiriou | 0.3205 | 1.055e-02 | 8.086e-03 | 1.943e-01 |
|---|
| Van-De-Vijver | 0.2945 | 7.220e-04 | 1.040e-03 | 4.712e-02 |
|---|
| Zhang | 0.2584 | 9.703e-02 | 5.852e-02 | 2.139e-01 |
|---|
| Zhou | 0.4385 | 4.522e-02 | 3.310e-02 | 2.904e-01 |
|---|
Expression data for subnetwork 1854-2-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
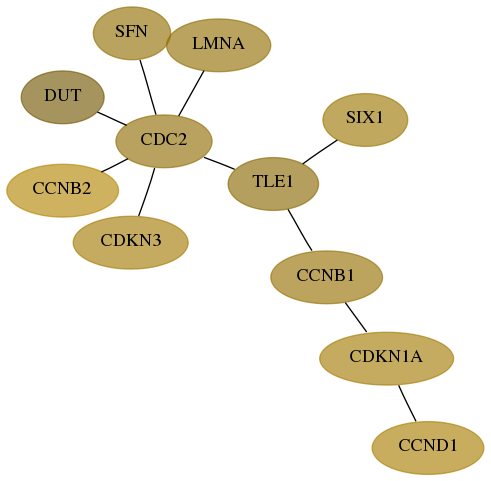
Score for each gene in subnetwork 1854-2-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| cdkn3 |   | 53 | 5 | 29 | 22 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.166 | -0.009 | 0.323 | 0.186 | 0.198 | 0.201 | 0.271 | 0.372 |
|---|
| dut |   | 1 | 116 | 182 | 188 | 0.036 | -0.023 | 0.106 | 0.211 | 0.114 | -0.048 | -0.296 | 0.003 | 0.119 | 0.177 | undef | 0.164 | -0.020 |
|---|
| ccnb2 |   | 12 | 18 | 84 | 62 | 0.164 | 0.176 | 0.204 | 0.123 | 0.172 | 0.178 | 0.090 | 0.366 | 0.240 | 0.169 | 0.279 | 0.211 | 0.463 |
|---|
| cdkn1a |   | 44 | 7 | 1 | 4 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | undef | -0.201 | 0.069 |
|---|
| tle1 |   | 47 | 6 | 1 | 3 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | 0.170 | 0.068 | 0.025 |
|---|
| lmna |   | 4 | 50 | 132 | 120 | 0.075 | 0.053 | 0.013 | -0.214 | 0.063 | 0.127 | 0.231 | 0.096 | -0.031 | 0.019 | 0.119 | -0.196 | 0.191 |
|---|
| ccnb1 |   | 42 | 8 | 56 | 39 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | undef | 0.167 | 0.398 |
|---|
| six1 |   | 13 | 17 | 123 | 96 | 0.097 | 0.114 | 0.095 | 0.149 | 0.151 | -0.015 | -0.006 | 0.123 | 0.113 | 0.184 | 0.082 | 0.191 | 0.079 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| sfn |   | 61 | 3 | 1 | 2 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.158 | 0.079 | 0.181 |
|---|
GO Enrichment output for subnetwork 1854-2-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.382E-09 | 1.008E-05 |
|---|
| deoxyribonucleoside metabolic process | GO:0009120 |  | 3.451E-08 | 3.969E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.315E-08 | 4.841E-05 |
|---|
| interphase | GO:0051325 |  | 7.092E-08 | 4.078E-05 |
|---|
| thymus development | GO:0048538 |  | 1.446E-07 | 6.651E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.247E-07 | 8.613E-05 |
|---|
| pyrimidine deoxyribonucleotide metabolic process | GO:0009219 |  | 2.835E-07 | 9.317E-05 |
|---|
| pyrimidine nucleoside triphosphate metabolic process | GO:0009147 |  | 3.778E-07 | 1.086E-04 |
|---|
| 2'-deoxyribonucleotide metabolic process | GO:0009394 |  | 1.163E-06 | 2.972E-04 |
|---|
| D-ribose metabolic process | GO:0006014 |  | 1.654E-06 | 3.805E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.267E-06 | 4.74E-04 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.022E-09 | 2.496E-06 |
|---|
| deoxyribonucleoside triphosphate metabolic process | GO:0009200 |  | 4.877E-09 | 5.958E-06 |
|---|
| deoxyribonucleoside metabolic process | GO:0009120 |  | 9.749E-09 | 7.939E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.619E-08 | 9.89E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.693E-08 | 2.781E-05 |
|---|
| thymus development | GO:0048538 |  | 8.021E-08 | 3.266E-05 |
|---|
| pyrimidine deoxyribonucleotide metabolic process | GO:0009219 |  | 1.069E-07 | 3.73E-05 |
|---|
| pyrimidine nucleoside triphosphate metabolic process | GO:0009147 |  | 1.389E-07 | 4.241E-05 |
|---|
| 2'-deoxyribonucleotide metabolic process | GO:0009394 |  | 4.69E-07 | 1.273E-04 |
|---|
| D-ribose metabolic process | GO:0006014 |  | 7.441E-07 | 1.818E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 8.553E-07 | 1.9E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.295E-09 | 3.115E-06 |
|---|
| deoxyribonucleoside triphosphate metabolic process | GO:0009200 |  | 5.705E-09 | 6.863E-06 |
|---|
| deoxyribonucleoside metabolic process | GO:0009120 |  | 1.14E-08 | 9.145E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.752E-08 | 1.054E-05 |
|---|
| interphase | GO:0051325 |  | 2.05E-08 | 9.866E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.946E-08 | 2.785E-05 |
|---|
| pyrimidine deoxyribonucleotide metabolic process | GO:0009219 |  | 9.381E-08 | 3.224E-05 |
|---|
| thymus development | GO:0048538 |  | 9.381E-08 | 2.821E-05 |
|---|
| pyrimidine nucleoside triphosphate metabolic process | GO:0009147 |  | 1.25E-07 | 3.342E-05 |
|---|
| 2'-deoxyribonucleotide metabolic process | GO:0009394 |  | 3.853E-07 | 9.269E-05 |
|---|
| D-ribose metabolic process | GO:0006014 |  | 6.448E-07 | 1.41E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.022E-09 | 2.496E-06 |
|---|
| deoxyribonucleoside triphosphate metabolic process | GO:0009200 |  | 4.877E-09 | 5.958E-06 |
|---|
| deoxyribonucleoside metabolic process | GO:0009120 |  | 9.749E-09 | 7.939E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.619E-08 | 9.89E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.693E-08 | 2.781E-05 |
|---|
| thymus development | GO:0048538 |  | 8.021E-08 | 3.266E-05 |
|---|
| pyrimidine deoxyribonucleotide metabolic process | GO:0009219 |  | 1.069E-07 | 3.73E-05 |
|---|
| pyrimidine nucleoside triphosphate metabolic process | GO:0009147 |  | 1.389E-07 | 4.241E-05 |
|---|
| 2'-deoxyribonucleotide metabolic process | GO:0009394 |  | 4.69E-07 | 1.273E-04 |
|---|
| D-ribose metabolic process | GO:0006014 |  | 7.441E-07 | 1.818E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 8.553E-07 | 1.9E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.295E-09 | 3.115E-06 |
|---|
| deoxyribonucleoside triphosphate metabolic process | GO:0009200 |  | 5.705E-09 | 6.863E-06 |
|---|
| deoxyribonucleoside metabolic process | GO:0009120 |  | 1.14E-08 | 9.145E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.752E-08 | 1.054E-05 |
|---|
| interphase | GO:0051325 |  | 2.05E-08 | 9.866E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.946E-08 | 2.785E-05 |
|---|
| pyrimidine deoxyribonucleotide metabolic process | GO:0009219 |  | 9.381E-08 | 3.224E-05 |
|---|
| thymus development | GO:0048538 |  | 9.381E-08 | 2.821E-05 |
|---|
| pyrimidine nucleoside triphosphate metabolic process | GO:0009147 |  | 1.25E-07 | 3.342E-05 |
|---|
| 2'-deoxyribonucleotide metabolic process | GO:0009394 |  | 3.853E-07 | 9.269E-05 |
|---|
| D-ribose metabolic process | GO:0006014 |  | 6.448E-07 | 1.41E-04 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.116E-11 | 3.255E-07 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.163E-09 | 2.077E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.259E-08 | 1.498E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.722E-08 | 1.537E-05 |
|---|
| thymus development | GO:0048538 |  | 3.704E-08 | 2.646E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 3.704E-08 | 2.205E-05 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 6.666E-08 | 3.401E-05 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 8.163E-08 | 3.644E-05 |
|---|
| kinase regulator activity | GO:0019207 |  | 1.414E-07 | 5.609E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.523E-07 | 5.44E-05 |
|---|
| protein kinase inhibitor activity | GO:0004860 |  | 7.31E-07 | 2.373E-04 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.599E-11 | 2.042E-07 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.058E-10 | 1.105E-06 |
|---|
| interphase | GO:0051325 |  | 6.861E-10 | 8.343E-07 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.869E-09 | 7.177E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 8.439E-09 | 6.157E-06 |
|---|
| thymus development | GO:0048538 |  | 3.311E-08 | 2.013E-05 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 4.564E-08 | 2.378E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 5.475E-08 | 2.496E-05 |
|---|
| kinase regulator activity | GO:0019207 |  | 8.1E-08 | 3.283E-05 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 8.417E-08 | 3.071E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 8.417E-08 | 2.791E-05 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.295E-09 | 3.115E-06 |
|---|
| deoxyribonucleoside triphosphate metabolic process | GO:0009200 |  | 5.705E-09 | 6.863E-06 |
|---|
| deoxyribonucleoside metabolic process | GO:0009120 |  | 1.14E-08 | 9.145E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.752E-08 | 1.054E-05 |
|---|
| interphase | GO:0051325 |  | 2.05E-08 | 9.866E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.946E-08 | 2.785E-05 |
|---|
| pyrimidine deoxyribonucleotide metabolic process | GO:0009219 |  | 9.381E-08 | 3.224E-05 |
|---|
| thymus development | GO:0048538 |  | 9.381E-08 | 2.821E-05 |
|---|
| pyrimidine nucleoside triphosphate metabolic process | GO:0009147 |  | 1.25E-07 | 3.342E-05 |
|---|
| 2'-deoxyribonucleotide metabolic process | GO:0009394 |  | 3.853E-07 | 9.269E-05 |
|---|
| D-ribose metabolic process | GO:0006014 |  | 6.448E-07 | 1.41E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.382E-09 | 1.008E-05 |
|---|
| deoxyribonucleoside metabolic process | GO:0009120 |  | 3.451E-08 | 3.969E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.315E-08 | 4.841E-05 |
|---|
| interphase | GO:0051325 |  | 7.092E-08 | 4.078E-05 |
|---|
| thymus development | GO:0048538 |  | 1.446E-07 | 6.651E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.247E-07 | 8.613E-05 |
|---|
| pyrimidine deoxyribonucleotide metabolic process | GO:0009219 |  | 2.835E-07 | 9.317E-05 |
|---|
| pyrimidine nucleoside triphosphate metabolic process | GO:0009147 |  | 3.778E-07 | 1.086E-04 |
|---|
| 2'-deoxyribonucleotide metabolic process | GO:0009394 |  | 1.163E-06 | 2.972E-04 |
|---|
| D-ribose metabolic process | GO:0006014 |  | 1.654E-06 | 3.805E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.267E-06 | 4.74E-04 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.382E-09 | 1.008E-05 |
|---|
| deoxyribonucleoside metabolic process | GO:0009120 |  | 3.451E-08 | 3.969E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.315E-08 | 4.841E-05 |
|---|
| interphase | GO:0051325 |  | 7.092E-08 | 4.078E-05 |
|---|
| thymus development | GO:0048538 |  | 1.446E-07 | 6.651E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.247E-07 | 8.613E-05 |
|---|
| pyrimidine deoxyribonucleotide metabolic process | GO:0009219 |  | 2.835E-07 | 9.317E-05 |
|---|
| pyrimidine nucleoside triphosphate metabolic process | GO:0009147 |  | 3.778E-07 | 1.086E-04 |
|---|
| 2'-deoxyribonucleotide metabolic process | GO:0009394 |  | 1.163E-06 | 2.972E-04 |
|---|
| D-ribose metabolic process | GO:0006014 |  | 1.654E-06 | 3.805E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.267E-06 | 4.74E-04 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| thymus development | GO:0048538 |  | 7.366E-09 | 1.357E-05 |
|---|
| muscle cell migration | GO:0014812 |  | 6.331E-06 | 5.834E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 9.493E-06 | 5.832E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 1.329E-05 | 6.121E-03 |
|---|
| induction of an organ | GO:0001759 |  | 1.329E-05 | 4.897E-03 |
|---|
| thyroid gland development | GO:0030878 |  | 1.771E-05 | 5.439E-03 |
|---|
| T cell homeostasis | GO:0043029 |  | 1.771E-05 | 4.662E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.771E-05 | 4.079E-03 |
|---|
| developmental induction | GO:0031128 |  | 1.771E-05 | 3.626E-03 |
|---|
| positive regulation of cell size | GO:0045793 |  | 2.276E-05 | 4.194E-03 |
|---|
| negative regulation of caspase activity | GO:0043154 |  | 2.276E-05 | 3.813E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.382E-09 | 1.008E-05 |
|---|
| deoxyribonucleoside metabolic process | GO:0009120 |  | 3.451E-08 | 3.969E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.315E-08 | 4.841E-05 |
|---|
| interphase | GO:0051325 |  | 7.092E-08 | 4.078E-05 |
|---|
| thymus development | GO:0048538 |  | 1.446E-07 | 6.651E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.247E-07 | 8.613E-05 |
|---|
| pyrimidine deoxyribonucleotide metabolic process | GO:0009219 |  | 2.835E-07 | 9.317E-05 |
|---|
| pyrimidine nucleoside triphosphate metabolic process | GO:0009147 |  | 3.778E-07 | 1.086E-04 |
|---|
| 2'-deoxyribonucleotide metabolic process | GO:0009394 |  | 1.163E-06 | 2.972E-04 |
|---|
| D-ribose metabolic process | GO:0006014 |  | 1.654E-06 | 3.805E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.267E-06 | 4.74E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.53E-09 | 1.075E-05 |
|---|
| deoxyribonucleoside metabolic process | GO:0009120 |  | 3.081E-08 | 3.655E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.677E-08 | 4.49E-05 |
|---|
| interphase | GO:0051325 |  | 6.367E-08 | 3.777E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.143E-07 | 1.017E-04 |
|---|
| thymus development | GO:0048538 |  | 2.532E-07 | 1.001E-04 |
|---|
| pyrimidine deoxyribonucleotide metabolic process | GO:0009219 |  | 3.373E-07 | 1.143E-04 |
|---|
| pyrimidine nucleoside triphosphate metabolic process | GO:0009147 |  | 4.381E-07 | 1.3E-04 |
|---|
| 2'-deoxyribonucleotide metabolic process | GO:0009394 |  | 1.245E-06 | 3.283E-04 |
|---|
| D-ribose metabolic process | GO:0006014 |  | 1.737E-06 | 4.121E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.024E-06 | 4.367E-04 |
|---|



