Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 116225-1-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1795 | 1.634e-02 | 1.306e-02 | 1.280e-01 |
|---|
| IPC-NIBC-129 | 0.2267 | 3.068e-02 | 4.506e-02 | 3.205e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2260 | 3.124e-02 | 4.697e-02 | 4.822e-01 |
|---|
| Loi_GPL570 | 0.2380 | 6.933e-02 | 7.818e-02 | 2.818e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1549 | 1.500e-05 | 1.240e-04 | 1.180e-04 |
|---|
| Parker_GPL1390 | 0.1868 | 5.232e-02 | 6.486e-02 | 4.038e-01 |
|---|
| Parker_GPL887 | 0.0514 | 4.074e-01 | 4.265e-01 | 4.510e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3128 | 7.939e-01 | 7.503e-01 | 9.986e-01 |
|---|
| Schmidt | 0.2398 | 3.199e-02 | 3.812e-02 | 3.180e-01 |
|---|
| Sotiriou | 0.3245 | 9.874e-03 | 7.549e-03 | 1.870e-01 |
|---|
| Van-De-Vijver | 0.2924 | 6.720e-04 | 9.670e-04 | 4.487e-02 |
|---|
| Zhang | 0.2420 | 1.449e-01 | 9.693e-02 | 2.990e-01 |
|---|
| Zhou | 0.4425 | 1.020e-01 | 9.517e-02 | 5.257e-01 |
|---|
Expression data for subnetwork 116225-1-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
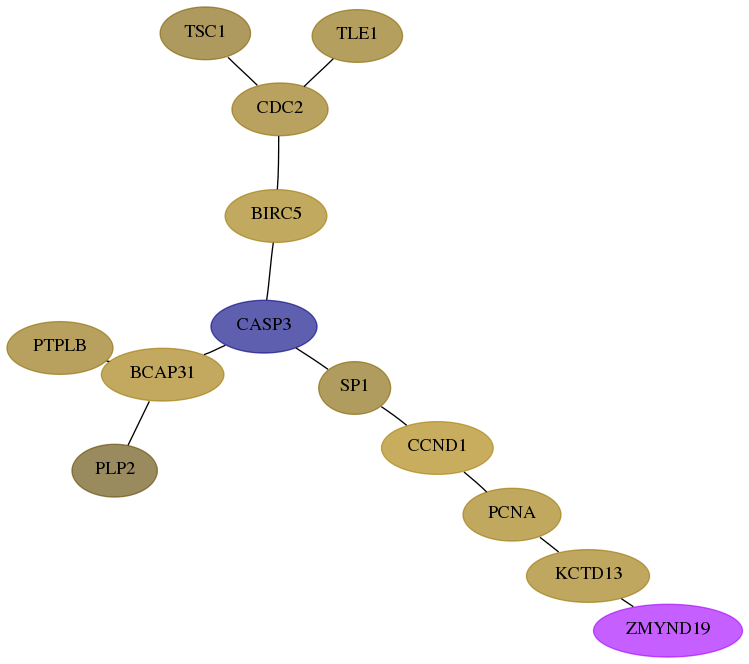
Score for each gene in subnetwork 116225-1-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| pcna |   | 15 | 11 | 110 | 85 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.119 | 0.104 | 0.256 | 0.151 | 0.059 | 0.201 | 0.123 | 0.233 |
|---|
| tle1 |   | 47 | 6 | 1 | 3 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | 0.170 | 0.068 | 0.025 |
|---|
| plp2 |   | 1 | 116 | 283 | 284 | 0.023 | 0.132 | 0.168 | 0.043 | 0.164 | 0.196 | 0.191 | 0.034 | 0.053 | 0.223 | 0.217 | -0.010 | 0.343 |
|---|
| ptplb |   | 1 | 116 | 283 | 284 | 0.070 | 0.038 | 0.194 | 0.139 | 0.169 | 0.101 | -0.438 | 0.132 | 0.168 | 0.180 | undef | 0.126 | 0.290 |
|---|
| kctd13 |   | 1 | 116 | 283 | 284 | 0.093 | 0.039 | 0.006 | 0.143 | 0.079 | -0.134 | 0.388 | 0.074 | -0.095 | 0.073 | undef | 0.055 | 0.304 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| casp3 |   | 2 | 78 | 103 | 102 | -0.051 | 0.158 | 0.146 | 0.160 | 0.074 | 0.210 | 0.252 | 0.202 | 0.070 | 0.073 | -0.058 | 0.059 | 0.305 |
|---|
| sp1 |   | 8 | 26 | 71 | 57 | 0.054 | 0.186 | 0.147 | 0.242 | 0.125 | 0.033 | -0.370 | 0.202 | 0.284 | 0.227 | 0.304 | 0.091 | -0.068 |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| zmynd19 |   | 1 | 116 | 283 | 284 | undef | 0.220 | 0.171 | 0.151 | 0.102 | -0.217 | 0.551 | 0.309 | undef | undef | undef | undef | undef |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| bcap31 |   | 1 | 116 | 283 | 284 | 0.103 | 0.079 | 0.104 | 0.144 | 0.177 | 0.184 | -0.267 | 0.132 | 0.105 | 0.198 | 0.145 | -0.086 | 0.180 |
|---|
| birc5 |   | 5 | 40 | 103 | 89 | 0.101 | 0.216 | 0.190 | 0.134 | 0.223 | 0.149 | 0.166 | 0.202 | 0.236 | 0.241 | undef | 0.205 | 0.400 |
|---|
GO Enrichment output for subnetwork 116225-1-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.708E-07 | 6.229E-04 |
|---|
| positive regulation of nuclear division | GO:0051785 |  | 3.908E-06 | 4.494E-03 |
|---|
| cytokinesis | GO:0000910 |  | 9.209E-06 | 7.06E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.508E-05 | 8.673E-03 |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 1.508E-05 | 6.938E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.508E-05 | 5.782E-03 |
|---|
| neuron apoptosis | GO:0051402 |  | 2.261E-05 | 7.428E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 2.261E-05 | 6.5E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 2.261E-05 | 5.777E-03 |
|---|
| negative regulation of B cell activation | GO:0050869 |  | 2.261E-05 | 5.2E-03 |
|---|
| response to UV | GO:0009411 |  | 2.535E-05 | 5.301E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.02E-08 | 1.226E-04 |
|---|
| positive regulation of nuclear division | GO:0051785 |  | 1.004E-06 | 1.227E-03 |
|---|
| cytokinesis | GO:0000910 |  | 4.399E-06 | 3.582E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.549E-06 | 3.389E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 5.549E-06 | 2.711E-03 |
|---|
| B cell homeostasis | GO:0001782 |  | 5.549E-06 | 2.259E-03 |
|---|
| neuron apoptosis | GO:0051402 |  | 8.319E-06 | 2.903E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 8.319E-06 | 2.541E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 8.319E-06 | 2.258E-03 |
|---|
| negative regulation of B cell activation | GO:0050869 |  | 8.319E-06 | 2.032E-03 |
|---|
| response to UV | GO:0009411 |  | 9.988E-06 | 2.218E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 9.699E-08 | 2.334E-04 |
|---|
| positive regulation of nuclear division | GO:0051785 |  | 1.648E-06 | 1.982E-03 |
|---|
| cytokinesis | GO:0000910 |  | 6.209E-06 | 4.98E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.727E-06 | 4.648E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 7.727E-06 | 3.718E-03 |
|---|
| B cell homeostasis | GO:0001782 |  | 7.727E-06 | 3.098E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 7.727E-06 | 2.656E-03 |
|---|
| neuron apoptosis | GO:0051402 |  | 1.158E-05 | 3.484E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.158E-05 | 3.097E-03 |
|---|
| negative regulation of B cell activation | GO:0050869 |  | 1.158E-05 | 2.787E-03 |
|---|
| response to UV | GO:0009411 |  | 1.461E-05 | 3.195E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.02E-08 | 1.226E-04 |
|---|
| positive regulation of nuclear division | GO:0051785 |  | 1.004E-06 | 1.227E-03 |
|---|
| cytokinesis | GO:0000910 |  | 4.399E-06 | 3.582E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.549E-06 | 3.389E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 5.549E-06 | 2.711E-03 |
|---|
| B cell homeostasis | GO:0001782 |  | 5.549E-06 | 2.259E-03 |
|---|
| neuron apoptosis | GO:0051402 |  | 8.319E-06 | 2.903E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 8.319E-06 | 2.541E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 8.319E-06 | 2.258E-03 |
|---|
| negative regulation of B cell activation | GO:0050869 |  | 8.319E-06 | 2.032E-03 |
|---|
| response to UV | GO:0009411 |  | 9.988E-06 | 2.218E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 9.699E-08 | 2.334E-04 |
|---|
| positive regulation of nuclear division | GO:0051785 |  | 1.648E-06 | 1.982E-03 |
|---|
| cytokinesis | GO:0000910 |  | 6.209E-06 | 4.98E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.727E-06 | 4.648E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 7.727E-06 | 3.718E-03 |
|---|
| B cell homeostasis | GO:0001782 |  | 7.727E-06 | 3.098E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 7.727E-06 | 2.656E-03 |
|---|
| neuron apoptosis | GO:0051402 |  | 1.158E-05 | 3.484E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.158E-05 | 3.097E-03 |
|---|
| negative regulation of B cell activation | GO:0050869 |  | 1.158E-05 | 2.787E-03 |
|---|
| response to UV | GO:0009411 |  | 1.461E-05 | 3.195E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.259E-08 | 4.494E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 3.704E-08 | 6.614E-05 |
|---|
| response to UV | GO:0009411 |  | 4.081E-06 | 4.857E-03 |
|---|
| embryonic camera-type eye development | GO:0031076 |  | 4.142E-06 | 3.697E-03 |
|---|
| MutLalpha complex binding | GO:0032405 |  | 4.142E-06 | 2.958E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 4.142E-06 | 2.465E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.339E-06 | 2.214E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.21E-06 | 2.772E-03 |
|---|
| enucleate erythrocyte differentiation | GO:0043353 |  | 6.21E-06 | 2.464E-03 |
|---|
| DNA replication factor C complex | GO:0005663 |  | 6.21E-06 | 2.218E-03 |
|---|
| B cell homeostasis | GO:0001782 |  | 6.21E-06 | 2.016E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.861E-08 | 6.787E-05 |
|---|
| midbody | GO:0030496 |  | 7.296E-08 | 1.331E-04 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.206E-07 | 1.467E-04 |
|---|
| spindle microtubule | GO:0005876 |  | 2.699E-07 | 2.462E-04 |
|---|
| chaperone binding | GO:0051087 |  | 3.768E-07 | 2.749E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.037E-06 | 2.455E-03 |
|---|
| response to UV | GO:0009411 |  | 4.638E-06 | 2.417E-03 |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 5.187E-06 | 2.365E-03 |
|---|
| enucleate erythrocyte differentiation | GO:0043353 |  | 5.187E-06 | 2.103E-03 |
|---|
| B cell homeostasis | GO:0001782 |  | 5.187E-06 | 1.892E-03 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 5.187E-06 | 1.72E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 9.699E-08 | 2.334E-04 |
|---|
| positive regulation of nuclear division | GO:0051785 |  | 1.648E-06 | 1.982E-03 |
|---|
| cytokinesis | GO:0000910 |  | 6.209E-06 | 4.98E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.727E-06 | 4.648E-03 |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 7.727E-06 | 3.718E-03 |
|---|
| B cell homeostasis | GO:0001782 |  | 7.727E-06 | 3.098E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 7.727E-06 | 2.656E-03 |
|---|
| neuron apoptosis | GO:0051402 |  | 1.158E-05 | 3.484E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.158E-05 | 3.097E-03 |
|---|
| negative regulation of B cell activation | GO:0050869 |  | 1.158E-05 | 2.787E-03 |
|---|
| response to UV | GO:0009411 |  | 1.461E-05 | 3.195E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.708E-07 | 6.229E-04 |
|---|
| positive regulation of nuclear division | GO:0051785 |  | 3.908E-06 | 4.494E-03 |
|---|
| cytokinesis | GO:0000910 |  | 9.209E-06 | 7.06E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.508E-05 | 8.673E-03 |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 1.508E-05 | 6.938E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.508E-05 | 5.782E-03 |
|---|
| neuron apoptosis | GO:0051402 |  | 2.261E-05 | 7.428E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 2.261E-05 | 6.5E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 2.261E-05 | 5.777E-03 |
|---|
| negative regulation of B cell activation | GO:0050869 |  | 2.261E-05 | 5.2E-03 |
|---|
| response to UV | GO:0009411 |  | 2.535E-05 | 5.301E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.708E-07 | 6.229E-04 |
|---|
| positive regulation of nuclear division | GO:0051785 |  | 3.908E-06 | 4.494E-03 |
|---|
| cytokinesis | GO:0000910 |  | 9.209E-06 | 7.06E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.508E-05 | 8.673E-03 |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 1.508E-05 | 6.938E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.508E-05 | 5.782E-03 |
|---|
| neuron apoptosis | GO:0051402 |  | 2.261E-05 | 7.428E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 2.261E-05 | 6.5E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 2.261E-05 | 5.777E-03 |
|---|
| negative regulation of B cell activation | GO:0050869 |  | 2.261E-05 | 5.2E-03 |
|---|
| response to UV | GO:0009411 |  | 2.535E-05 | 5.301E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 1.771E-05 | 0.03263475 |
|---|
| regulation of activated T cell proliferation | GO:0046006 |  | 1.771E-05 | 0.01631737 |
|---|
| negative regulation of T cell proliferation | GO:0042130 |  | 1.771E-05 | 0.01087825 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.771E-05 | 8.159E-03 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 1.771E-05 | 6.527E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 1.771E-05 | 5.439E-03 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 2.478E-05 | 6.523E-03 |
|---|
| negative regulation of lymphocyte proliferation | GO:0050672 |  | 2.478E-05 | 5.708E-03 |
|---|
| negative regulation of T cell activation | GO:0050868 |  | 2.478E-05 | 5.074E-03 |
|---|
| regulation of stress fiber formation | GO:0051492 |  | 3.302E-05 | 6.085E-03 |
|---|
| T cell homeostasis | GO:0043029 |  | 3.302E-05 | 5.532E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.708E-07 | 6.229E-04 |
|---|
| positive regulation of nuclear division | GO:0051785 |  | 3.908E-06 | 4.494E-03 |
|---|
| cytokinesis | GO:0000910 |  | 9.209E-06 | 7.06E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.508E-05 | 8.673E-03 |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 1.508E-05 | 6.938E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.508E-05 | 5.782E-03 |
|---|
| neuron apoptosis | GO:0051402 |  | 2.261E-05 | 7.428E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 2.261E-05 | 6.5E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 2.261E-05 | 5.777E-03 |
|---|
| negative regulation of B cell activation | GO:0050869 |  | 2.261E-05 | 5.2E-03 |
|---|
| response to UV | GO:0009411 |  | 2.535E-05 | 5.301E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.06E-05 | 0.02515107 |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 1.06E-05 | 0.01257554 |
|---|
| B cell homeostasis | GO:0001782 |  | 1.06E-05 | 8.384E-03 |
|---|
| definitive hemopoiesis | GO:0060216 |  | 1.06E-05 | 6.288E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.399E-05 | 6.641E-03 |
|---|
| embryonic camera-type eye development | GO:0031076 |  | 1.589E-05 | 6.284E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.589E-05 | 5.386E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 1.589E-05 | 4.713E-03 |
|---|
| response to UV | GO:0009411 |  | 1.597E-05 | 4.21E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.223E-05 | 5.275E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 2.223E-05 | 4.795E-03 |
|---|



