Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 10808-0-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2472 | 5.921e-03 | 4.128e-03 | 5.705e-02 |
|---|
| IPC-NIBC-129 | 0.2893 | 2.429e-02 | 3.598e-02 | 2.700e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2752 | 5.092e-03 | 8.978e-03 | 1.850e-01 |
|---|
| Loi_GPL570 | 0.2432 | 6.443e-02 | 7.280e-02 | 2.664e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1557 | 0.000e+00 | 3.000e-06 | 0.000e+00 |
|---|
| Parker_GPL1390 | 0.2708 | 6.280e-02 | 7.717e-02 | 4.451e-01 |
|---|
| Parker_GPL887 | 0.1920 | 2.682e-01 | 2.855e-01 | 2.792e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3485 | 2.235e-01 | 1.623e-01 | 7.858e-01 |
|---|
| Schmidt | 0.2270 | 8.238e-02 | 1.027e-01 | 5.452e-01 |
|---|
| Sotiriou | 0.3040 | 1.391e-02 | 1.075e-02 | 2.270e-01 |
|---|
| Van-De-Vijver | 0.3092 | 1.589e-03 | 2.285e-03 | 7.969e-02 |
|---|
| Zhang | 0.1833 | 1.432e-01 | 9.554e-02 | 2.962e-01 |
|---|
| Zhou | 0.3944 | 7.385e-02 | 6.289e-02 | 4.226e-01 |
|---|
Expression data for subnetwork 10808-0-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
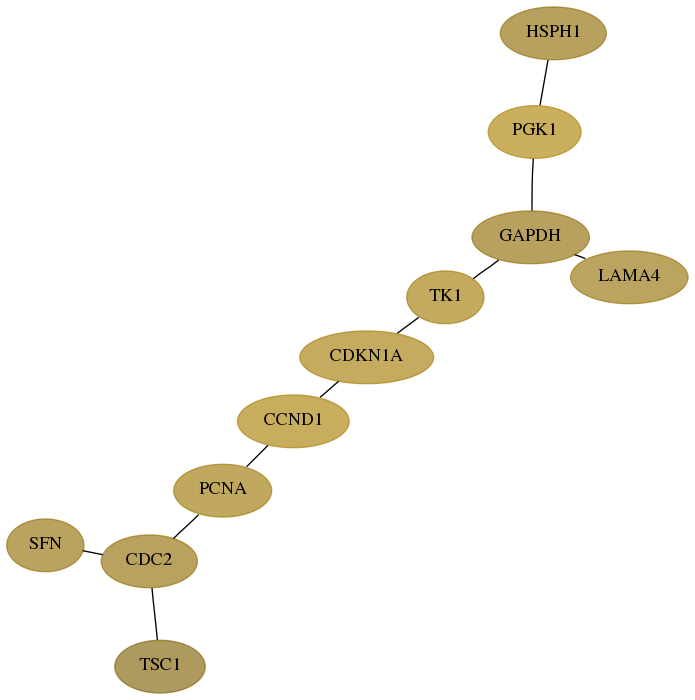
Score for each gene in subnetwork 10808-0-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| pcna |   | 15 | 11 | 110 | 85 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.119 | 0.104 | 0.256 | 0.151 | 0.059 | 0.201 | 0.123 | 0.233 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| tk1 |   | 7 | 30 | 84 | 67 | 0.109 | 0.149 | 0.190 | -0.017 | 0.131 | 0.105 | -0.100 | 0.257 | 0.204 | 0.142 | 0.293 | 0.164 | 0.222 |
|---|
| gapdh |   | 6 | 35 | 18 | 20 | 0.071 | 0.312 | 0.102 | 0.145 | 0.165 | 0.172 | -0.253 | 0.288 | 0.064 | 0.213 | 0.225 | -0.088 | 0.235 |
|---|
| cdkn1a |   | 44 | 7 | 1 | 4 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | undef | -0.201 | 0.069 |
|---|
| hsph1 |   | 8 | 26 | 18 | 16 | 0.071 | 0.260 | 0.138 | 0.050 | 0.148 | 0.197 | -0.116 | 0.161 | 0.143 | 0.132 | -0.006 | 0.094 | 0.357 |
|---|
| sfn |   | 61 | 3 | 1 | 2 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.158 | 0.079 | 0.181 |
|---|
| pgk1 |   | 5 | 40 | 18 | 23 | 0.141 | 0.293 | 0.135 | 0.197 | 0.170 | 0.170 | 0.120 | 0.310 | 0.201 | 0.127 | -0.046 | 0.054 | 0.141 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| lama4 |   | 6 | 35 | 84 | 73 | 0.079 | 0.190 | 0.023 | -0.039 | 0.041 | 0.126 | -0.167 | -0.168 | -0.007 | -0.089 | 0.028 | 0.057 | 0.089 |
|---|
GO Enrichment output for subnetwork 10808-0-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 1.29E-11 | 2.968E-08 |
|---|
| glucose catabolic process | GO:0006007 |  | 4.685E-11 | 5.387E-08 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.349E-10 | 1.034E-07 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 2.563E-10 | 1.474E-07 |
|---|
| alcohol catabolic process | GO:0046164 |  | 5.34E-10 | 2.456E-07 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.341E-09 | 5.14E-07 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.727E-08 | 5.674E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.446E-07 | 4.157E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.52E-07 | 1.155E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.508E-05 | 3.469E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.508E-05 | 3.154E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 1.234E-12 | 3.015E-09 |
|---|
| glucose catabolic process | GO:0006007 |  | 4.255E-12 | 5.197E-09 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.425E-11 | 1.161E-08 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 3.073E-11 | 1.877E-08 |
|---|
| alcohol catabolic process | GO:0046164 |  | 6.51E-11 | 3.181E-08 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 4.373E-09 | 1.781E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 7.193E-08 | 2.51E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.182E-07 | 3.609E-05 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.975E-06 | 1.622E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.076E-06 | 1.484E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.496E-06 | 1.443E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 1.363E-12 | 3.28E-09 |
|---|
| glucose catabolic process | GO:0006007 |  | 4.826E-12 | 5.806E-09 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.651E-11 | 1.324E-08 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 3.599E-11 | 2.165E-08 |
|---|
| alcohol catabolic process | GO:0046164 |  | 7.697E-11 | 3.704E-08 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 5.047E-09 | 2.024E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 8.3E-08 | 2.853E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.415E-07 | 4.257E-05 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.605E-06 | 1.766E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 6.605E-06 | 1.589E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.006E-06 | 1.532E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 1.234E-12 | 3.015E-09 |
|---|
| glucose catabolic process | GO:0006007 |  | 4.255E-12 | 5.197E-09 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.425E-11 | 1.161E-08 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 3.073E-11 | 1.877E-08 |
|---|
| alcohol catabolic process | GO:0046164 |  | 6.51E-11 | 3.181E-08 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 4.373E-09 | 1.781E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 7.193E-08 | 2.51E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.182E-07 | 3.609E-05 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.975E-06 | 1.622E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.076E-06 | 1.484E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.496E-06 | 1.443E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 1.363E-12 | 3.28E-09 |
|---|
| glucose catabolic process | GO:0006007 |  | 4.826E-12 | 5.806E-09 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.651E-11 | 1.324E-08 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 3.599E-11 | 2.165E-08 |
|---|
| alcohol catabolic process | GO:0046164 |  | 7.697E-11 | 3.704E-08 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 5.047E-09 | 2.024E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 8.3E-08 | 2.853E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.415E-07 | 4.257E-05 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.605E-06 | 1.766E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 6.605E-06 | 1.589E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.006E-06 | 1.532E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 7.479E-12 | 2.671E-08 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.427E-08 | 4.334E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 3.704E-08 | 4.409E-05 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 6.666E-08 | 5.951E-05 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 8.163E-08 | 5.83E-05 |
|---|
| kinase regulator activity | GO:0019207 |  | 1.414E-07 | 8.414E-05 |
|---|
| protein kinase inhibitor activity | GO:0004860 |  | 7.31E-07 | 3.729E-04 |
|---|
| kinase inhibitor activity | GO:0019210 |  | 9.053E-07 | 4.041E-04 |
|---|
| glycolysis | GO:0006096 |  | 2.549E-06 | 1.011E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.739E-06 | 9.781E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.366E-06 | 1.093E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 4.51E-12 | 1.645E-08 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.59E-08 | 2.899E-05 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 4.564E-08 | 5.55E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 5.475E-08 | 4.993E-05 |
|---|
| kinase regulator activity | GO:0019207 |  | 8.1E-08 | 5.91E-05 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 8.417E-08 | 5.118E-05 |
|---|
| protein kinase inhibitor activity | GO:0004860 |  | 8.943E-07 | 4.66E-04 |
|---|
| kinase inhibitor activity | GO:0019210 |  | 9.778E-07 | 4.459E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.589E-06 | 6.443E-04 |
|---|
| glycolysis | GO:0006096 |  | 1.589E-06 | 5.798E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.839E-06 | 6.099E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 1.363E-12 | 3.28E-09 |
|---|
| glucose catabolic process | GO:0006007 |  | 4.826E-12 | 5.806E-09 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.651E-11 | 1.324E-08 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 3.599E-11 | 2.165E-08 |
|---|
| alcohol catabolic process | GO:0046164 |  | 7.697E-11 | 3.704E-08 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 5.047E-09 | 2.024E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 8.3E-08 | 2.853E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.415E-07 | 4.257E-05 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.605E-06 | 1.766E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 6.605E-06 | 1.589E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.006E-06 | 1.532E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 1.29E-11 | 2.968E-08 |
|---|
| glucose catabolic process | GO:0006007 |  | 4.685E-11 | 5.387E-08 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.349E-10 | 1.034E-07 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 2.563E-10 | 1.474E-07 |
|---|
| alcohol catabolic process | GO:0046164 |  | 5.34E-10 | 2.456E-07 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.341E-09 | 5.14E-07 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.727E-08 | 5.674E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.446E-07 | 4.157E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.52E-07 | 1.155E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.508E-05 | 3.469E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.508E-05 | 3.154E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 1.29E-11 | 2.968E-08 |
|---|
| glucose catabolic process | GO:0006007 |  | 4.685E-11 | 5.387E-08 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.349E-10 | 1.034E-07 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 2.563E-10 | 1.474E-07 |
|---|
| alcohol catabolic process | GO:0046164 |  | 5.34E-10 | 2.456E-07 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.341E-09 | 5.14E-07 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.727E-08 | 5.674E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.446E-07 | 4.157E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.52E-07 | 1.155E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.508E-05 | 3.469E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.508E-05 | 3.154E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 3.965E-06 | 7.308E-03 |
|---|
| glucose catabolic process | GO:0006007 |  | 7.535E-06 | 6.944E-03 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.277E-05 | 7.842E-03 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.518E-05 | 6.995E-03 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 1.61E-05 | 5.934E-03 |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.861E-05 | 5.717E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 2.276E-05 | 5.992E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.276E-05 | 5.243E-03 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 2.276E-05 | 4.66E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 2.276E-05 | 4.194E-03 |
|---|
| regulation of embryonic development | GO:0045995 |  | 2.276E-05 | 3.813E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 1.29E-11 | 2.968E-08 |
|---|
| glucose catabolic process | GO:0006007 |  | 4.685E-11 | 5.387E-08 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.349E-10 | 1.034E-07 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 2.563E-10 | 1.474E-07 |
|---|
| alcohol catabolic process | GO:0046164 |  | 5.34E-10 | 2.456E-07 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.341E-09 | 5.14E-07 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.727E-08 | 5.674E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.446E-07 | 4.157E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.52E-07 | 1.155E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.508E-05 | 3.469E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.508E-05 | 3.154E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 2.237E-13 | 5.307E-10 |
|---|
| glucose catabolic process | GO:0006007 |  | 9.963E-13 | 1.182E-09 |
|---|
| hexose catabolic process | GO:0019320 |  | 3.067E-12 | 2.426E-09 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 6.116E-12 | 3.628E-09 |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.356E-11 | 6.437E-09 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 3.993E-11 | 1.579E-08 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.98E-08 | 6.712E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 2.366E-07 | 7.017E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.433E-07 | 1.696E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.639E-05 | 3.89E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.201E-05 | 4.749E-03 |
|---|



