Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1033-0-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2041 | 1.495e-02 | 1.182e-02 | 1.196e-01 |
|---|
| IPC-NIBC-129 | 0.2604 | 3.214e-02 | 4.712e-02 | 3.313e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2303 | 1.841e-02 | 2.902e-02 | 3.753e-01 |
|---|
| Loi_GPL570 | 0.1926 | 1.299e-01 | 1.438e-01 | 4.426e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1829 | 1.000e-06 | 1.600e-05 | 2.000e-06 |
|---|
| Parker_GPL1390 | 0.2116 | 7.207e-02 | 8.797e-02 | 4.778e-01 |
|---|
| Parker_GPL887 | 0.0995 | 3.507e-01 | 3.694e-01 | 3.810e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.4078 | 5.437e-01 | 4.728e-01 | 9.764e-01 |
|---|
| Schmidt | 0.2173 | 2.148e-02 | 2.497e-02 | 2.454e-01 |
|---|
| Sotiriou | 0.2970 | 1.561e-02 | 1.210e-02 | 2.419e-01 |
|---|
| Van-De-Vijver | 0.2890 | 8.860e-04 | 1.276e-03 | 5.412e-02 |
|---|
| Zhang | 0.2664 | 9.495e-02 | 5.695e-02 | 2.101e-01 |
|---|
| Zhou | 0.4271 | 4.280e-02 | 3.077e-02 | 2.775e-01 |
|---|
Expression data for subnetwork 1033-0-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
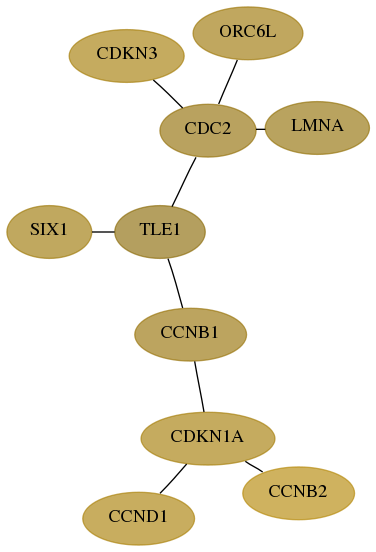
Score for each gene in subnetwork 1033-0-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| cdkn3 |   | 53 | 5 | 29 | 22 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.166 | -0.009 | 0.323 | 0.186 | 0.198 | 0.201 | 0.271 | 0.372 |
|---|
| ccnb2 |   | 12 | 18 | 84 | 62 | 0.164 | 0.176 | 0.204 | 0.123 | 0.172 | 0.178 | 0.090 | 0.366 | 0.240 | 0.169 | 0.279 | 0.211 | 0.463 |
|---|
| cdkn1a |   | 44 | 7 | 1 | 4 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | undef | -0.201 | 0.069 |
|---|
| tle1 |   | 47 | 6 | 1 | 3 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | 0.170 | 0.068 | 0.025 |
|---|
| lmna |   | 4 | 50 | 132 | 120 | 0.075 | 0.053 | 0.013 | -0.214 | 0.063 | 0.127 | 0.231 | 0.096 | -0.031 | 0.019 | 0.119 | -0.196 | 0.191 |
|---|
| ccnb1 |   | 42 | 8 | 56 | 39 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | undef | 0.167 | 0.398 |
|---|
| six1 |   | 13 | 17 | 123 | 96 | 0.097 | 0.114 | 0.095 | 0.149 | 0.151 | -0.015 | -0.006 | 0.123 | 0.113 | 0.184 | 0.082 | 0.191 | 0.079 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| orc6l |   | 15 | 11 | 34 | 28 | 0.094 | 0.270 | 0.084 | 0.114 | 0.106 | 0.240 | 0.256 | 0.234 | 0.192 | 0.105 | 0.258 | 0.175 | 0.160 |
|---|
GO Enrichment output for subnetwork 1033-0-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.821E-08 | 6.488E-05 |
|---|
| interphase | GO:0051325 |  | 3.169E-08 | 3.644E-05 |
|---|
| thymus development | GO:0048538 |  | 9.105E-08 | 6.98E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.197E-07 | 6.883E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.411E-07 | 1.109E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.43E-06 | 5.481E-04 |
|---|
| response to UV | GO:0009411 |  | 1.604E-05 | 5.271E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 6.735E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.121E-05 | 7.977E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 3.121E-05 | 7.179E-03 |
|---|
| induction of an organ | GO:0001759 |  | 3.121E-05 | 6.526E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 9.028E-09 | 2.206E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.593E-08 | 4.389E-05 |
|---|
| thymus development | GO:0048538 |  | 5.71E-08 | 4.65E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.651E-08 | 5.284E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 7.906E-07 | 3.863E-04 |
|---|
| muscle cell migration | GO:0014812 |  | 5.138E-06 | 2.092E-03 |
|---|
| response to UV | GO:0009411 |  | 8.888E-06 | 3.102E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.437E-05 | 4.387E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.687E-05 | 4.58E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.763E-05 | 4.307E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 2.004E-05 | 4.45E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 9.031E-09 | 2.173E-05 |
|---|
| interphase | GO:0051325 |  | 1.057E-08 | 1.271E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.128E-08 | 3.31E-05 |
|---|
| thymus development | GO:0048538 |  | 6.393E-08 | 3.846E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.936E-08 | 4.781E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 8.85E-07 | 3.549E-04 |
|---|
| muscle cell migration | GO:0014812 |  | 5.571E-06 | 1.915E-03 |
|---|
| response to UV | GO:0009411 |  | 8.889E-06 | 2.673E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.558E-05 | 4.164E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.888E-05 | 4.541E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.972E-05 | 4.314E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 9.028E-09 | 2.206E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.593E-08 | 4.389E-05 |
|---|
| thymus development | GO:0048538 |  | 5.71E-08 | 4.65E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.651E-08 | 5.284E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 7.906E-07 | 3.863E-04 |
|---|
| muscle cell migration | GO:0014812 |  | 5.138E-06 | 2.092E-03 |
|---|
| response to UV | GO:0009411 |  | 8.888E-06 | 3.102E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.437E-05 | 4.387E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.687E-05 | 4.58E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.763E-05 | 4.307E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 2.004E-05 | 4.45E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 9.031E-09 | 2.173E-05 |
|---|
| interphase | GO:0051325 |  | 1.057E-08 | 1.271E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.128E-08 | 3.31E-05 |
|---|
| thymus development | GO:0048538 |  | 6.393E-08 | 3.846E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.936E-08 | 4.781E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 8.85E-07 | 3.549E-04 |
|---|
| muscle cell migration | GO:0014812 |  | 5.571E-06 | 1.915E-03 |
|---|
| response to UV | GO:0009411 |  | 8.889E-06 | 2.673E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.558E-05 | 4.164E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.888E-05 | 4.541E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.972E-05 | 4.314E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.842E-10 | 2.086E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 8.812E-09 | 1.573E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.036E-08 | 1.233E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.46E-08 | 1.303E-05 |
|---|
| thymus development | GO:0048538 |  | 2.594E-08 | 1.853E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 2.594E-08 | 1.544E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.067E-07 | 5.444E-05 |
|---|
| response to UV | GO:0009411 |  | 2.863E-06 | 1.278E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.969E-06 | 1.972E-03 |
|---|
| nuclear origin of replication recognition complex | GO:0005664 |  | 4.969E-06 | 1.774E-03 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 6.445E-06 | 2.092E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.315E-10 | 1.209E-06 |
|---|
| interphase | GO:0051325 |  | 3.755E-10 | 6.85E-07 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.016E-09 | 6.099E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 6.139E-09 | 5.598E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.014E-08 | 7.395E-06 |
|---|
| thymus development | GO:0048538 |  | 2.409E-08 | 1.465E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 3.983E-08 | 2.076E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 6.125E-08 | 2.793E-05 |
|---|
| response to UV | GO:0009411 |  | 1.539E-06 | 6.24E-04 |
|---|
| origin recognition complex | GO:0000808 |  | 2.566E-06 | 9.361E-04 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 2.566E-06 | 8.51E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 9.031E-09 | 2.173E-05 |
|---|
| interphase | GO:0051325 |  | 1.057E-08 | 1.271E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.128E-08 | 3.31E-05 |
|---|
| thymus development | GO:0048538 |  | 6.393E-08 | 3.846E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.936E-08 | 4.781E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 8.85E-07 | 3.549E-04 |
|---|
| muscle cell migration | GO:0014812 |  | 5.571E-06 | 1.915E-03 |
|---|
| response to UV | GO:0009411 |  | 8.889E-06 | 2.673E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.558E-05 | 4.164E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.888E-05 | 4.541E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.972E-05 | 4.314E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.821E-08 | 6.488E-05 |
|---|
| interphase | GO:0051325 |  | 3.169E-08 | 3.644E-05 |
|---|
| thymus development | GO:0048538 |  | 9.105E-08 | 6.98E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.197E-07 | 6.883E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.411E-07 | 1.109E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.43E-06 | 5.481E-04 |
|---|
| response to UV | GO:0009411 |  | 1.604E-05 | 5.271E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 6.735E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.121E-05 | 7.977E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 3.121E-05 | 7.179E-03 |
|---|
| induction of an organ | GO:0001759 |  | 3.121E-05 | 6.526E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.821E-08 | 6.488E-05 |
|---|
| interphase | GO:0051325 |  | 3.169E-08 | 3.644E-05 |
|---|
| thymus development | GO:0048538 |  | 9.105E-08 | 6.98E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.197E-07 | 6.883E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.411E-07 | 1.109E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.43E-06 | 5.481E-04 |
|---|
| response to UV | GO:0009411 |  | 1.604E-05 | 5.271E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 6.735E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.121E-05 | 7.977E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 3.121E-05 | 7.179E-03 |
|---|
| induction of an organ | GO:0001759 |  | 3.121E-05 | 6.526E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| thymus development | GO:0048538 |  | 7.366E-09 | 1.357E-05 |
|---|
| muscle cell migration | GO:0014812 |  | 6.331E-06 | 5.834E-03 |
|---|
| induction of an organ | GO:0001759 |  | 1.329E-05 | 8.162E-03 |
|---|
| thyroid gland development | GO:0030878 |  | 1.771E-05 | 8.159E-03 |
|---|
| T cell homeostasis | GO:0043029 |  | 1.771E-05 | 6.527E-03 |
|---|
| developmental induction | GO:0031128 |  | 1.771E-05 | 5.439E-03 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 2.276E-05 | 5.992E-03 |
|---|
| lymphocyte homeostasis | GO:0002260 |  | 2.844E-05 | 6.551E-03 |
|---|
| ureteric bud development | GO:0001657 |  | 4.923E-05 | 0.01008166 |
|---|
| negative regulation of Wnt receptor signaling pathway | GO:0030178 |  | 4.923E-05 | 9.073E-03 |
|---|
| leukocyte homeostasis | GO:0001776 |  | 6.622E-05 | 0.01109531 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.821E-08 | 6.488E-05 |
|---|
| interphase | GO:0051325 |  | 3.169E-08 | 3.644E-05 |
|---|
| thymus development | GO:0048538 |  | 9.105E-08 | 6.98E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.197E-07 | 6.883E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.411E-07 | 1.109E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.43E-06 | 5.481E-04 |
|---|
| response to UV | GO:0009411 |  | 1.604E-05 | 5.271E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 6.735E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.121E-05 | 7.977E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 3.121E-05 | 7.179E-03 |
|---|
| induction of an organ | GO:0001759 |  | 3.121E-05 | 6.526E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.58E-08 | 3.749E-05 |
|---|
| interphase | GO:0051325 |  | 1.773E-08 | 2.103E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.882E-08 | 6.234E-05 |
|---|
| thymus development | GO:0048538 |  | 1.214E-07 | 7.201E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.691E-07 | 8.025E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 9.725E-07 | 3.846E-04 |
|---|
| response to UV | GO:0009411 |  | 1.167E-05 | 3.957E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.811E-05 | 5.371E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.413E-05 | 6.362E-03 |
|---|
| induction of an organ | GO:0001759 |  | 3.101E-05 | 7.358E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.256E-05 | 7.025E-03 |
|---|



