Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 9219-10-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1624 | 1.584e-02 | 1.898e-02 | 1.390e-01 |
|---|
| IPC-NIBC-129 | 0.2676 | 2.937e-02 | 3.602e-02 | 3.085e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2259 | 4.763e-03 | 7.900e-03 | 1.651e-01 |
|---|
| Loi_GPL570 | 0.2776 | 4.829e-02 | 4.793e-02 | 2.484e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1633 | 7.000e-06 | 4.700e-05 | 4.300e-05 |
|---|
| Parker_GPL1390 | 0.2967 | 1.647e-01 | 1.708e-01 | 6.762e-01 |
|---|
| Parker_GPL887 | 0.0421 | 8.711e-01 | 8.844e-01 | 9.173e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3307 | 1.126e-01 | 8.395e-02 | 5.562e-01 |
|---|
| Schmidt | 0.2444 | 6.415e-03 | 9.339e-03 | 9.209e-02 |
|---|
| Sotiriou | 0.3422 | 1.484e-03 | 3.740e-04 | 4.254e-02 |
|---|
| Wang | 0.1698 | 1.050e-01 | 7.281e-02 | 3.773e-01 |
|---|
| Zhang | 0.2596 | 1.212e-01 | 8.241e-02 | 2.842e-01 |
|---|
| Zhou | 0.4442 | 8.098e-02 | 8.784e-02 | 4.738e-01 |
|---|
Expression data for subnetwork 9219-10-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
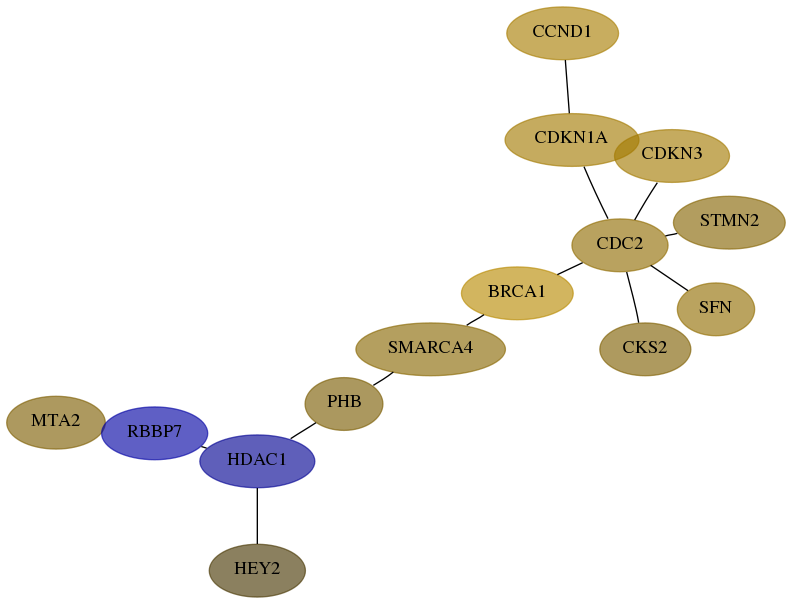
Score for each gene in subnetwork 9219-10-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| cdkn3 |   | 15 | 15 | 128 | 104 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.166 | -0.009 | 0.323 | 0.186 | 0.198 | 0.134 | 0.271 | 0.372 |
|---|
| hdac1 |   | 2 | 104 | 219 | 213 | -0.076 | 0.003 | -0.041 | 0.108 | 0.111 | 0.128 | 0.164 | -0.034 | -0.074 | 0.152 | -0.019 | 0.086 | 0.292 |
|---|
| stmn2 |   | 29 | 8 | 1 | 3 | 0.057 | 0.106 | 0.005 | 0.052 | 0.110 | 0.071 | -0.099 | 0.006 | 0.034 | 0.103 | 0.075 | 0.113 | -0.039 |
|---|
| mta2 |   | 1 | 164 | 219 | 230 | 0.047 | 0.113 | 0.051 | 0.096 | 0.048 | 0.256 | 0.143 | 0.086 | 0.230 | 0.033 | 0.025 | -0.039 | -0.052 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| hey2 |   | 2 | 104 | 219 | 213 | 0.013 | 0.189 | -0.017 | 0.127 | 0.076 | 0.165 | -0.287 | -0.039 | 0.152 | 0.076 | 0.057 | -0.043 | 0.367 |
|---|
| brca1 |   | 8 | 33 | 175 | 150 | 0.192 | 0.052 | 0.112 | 0.022 | 0.125 | -0.124 | 0.305 | 0.113 | 0.077 | 0.168 | 0.155 | 0.064 | 0.209 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| cks2 |   | 44 | 6 | 73 | 54 | 0.050 | 0.233 | 0.170 | 0.103 | 0.186 | 0.172 | 0.093 | 0.167 | 0.263 | 0.151 | 0.145 | 0.284 | 0.383 |
|---|
| rbbp7 |   | 3 | 79 | 219 | 206 | -0.113 | 0.089 | 0.051 | 0.076 | 0.106 | -0.053 | 0.092 | -0.020 | 0.126 | 0.138 | 0.115 | 0.168 | 0.280 |
|---|
| smarca4 |   | 1 | 164 | 219 | 230 | 0.062 | 0.223 | 0.136 | 0.131 | 0.107 | 0.119 | -0.044 | 0.222 | 0.120 | 0.093 | 0.000 | 0.008 | 0.281 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| phb |   | 1 | 164 | 219 | 230 | 0.044 | 0.165 | 0.102 | 0.115 | 0.161 | -0.003 | -0.122 | 0.301 | 0.098 | 0.145 | -0.015 | 0.082 | 0.212 |
|---|
| sfn |   | 50 | 4 | 29 | 17 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.025 | 0.079 | 0.181 |
|---|
GO Enrichment output for subnetwork 9219-10-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.72E-11 | 1.776E-07 |
|---|
| histone deacetylation | GO:0016575 |  | 1.066E-09 | 1.226E-06 |
|---|
| covalent chromatin modification | GO:0016569 |  | 5.757E-09 | 4.414E-06 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 5.85E-09 | 3.364E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.87E-07 | 1.32E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.865E-07 | 1.482E-04 |
|---|
| histone modification | GO:0016570 |  | 5.096E-07 | 1.674E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 4.489E-06 | 1.291E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.381E-06 | 1.631E-03 |
|---|
| interphase | GO:0051325 |  | 6.994E-06 | 1.609E-03 |
|---|
| keratinocyte differentiation | GO:0030216 |  | 1.64E-05 | 3.43E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.986E-12 | 1.462E-08 |
|---|
| histone deacetylation | GO:0016575 |  | 4.846E-10 | 5.919E-07 |
|---|
| covalent chromatin modification | GO:0016569 |  | 7.73E-10 | 6.295E-07 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 1.715E-09 | 1.047E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 5.201E-08 | 2.541E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.693E-08 | 2.318E-05 |
|---|
| histone modification | GO:0016570 |  | 9.412E-08 | 3.285E-05 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 1.11E-06 | 3.388E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.224E-06 | 3.322E-04 |
|---|
| glial cell fate commitment | GO:0021781 |  | 6.418E-06 | 1.568E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 6.418E-06 | 1.425E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.006E-11 | 2.42E-08 |
|---|
| histone deacetylation | GO:0016575 |  | 6.943E-10 | 8.352E-07 |
|---|
| covalent chromatin modification | GO:0016569 |  | 9.878E-10 | 7.922E-07 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 2.457E-09 | 1.478E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.441E-08 | 3.58E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.144E-08 | 3.266E-05 |
|---|
| histone modification | GO:0016570 |  | 1.147E-07 | 3.941E-05 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 1.458E-06 | 4.386E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.542E-06 | 4.122E-04 |
|---|
| interphase | GO:0051325 |  | 1.746E-06 | 4.202E-04 |
|---|
| keratinocyte differentiation | GO:0030216 |  | 6.695E-06 | 1.464E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.986E-12 | 1.462E-08 |
|---|
| histone deacetylation | GO:0016575 |  | 4.846E-10 | 5.919E-07 |
|---|
| covalent chromatin modification | GO:0016569 |  | 7.73E-10 | 6.295E-07 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 1.715E-09 | 1.047E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 5.201E-08 | 2.541E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.693E-08 | 2.318E-05 |
|---|
| histone modification | GO:0016570 |  | 9.412E-08 | 3.285E-05 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 1.11E-06 | 3.388E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.224E-06 | 3.322E-04 |
|---|
| glial cell fate commitment | GO:0021781 |  | 6.418E-06 | 1.568E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 6.418E-06 | 1.425E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.006E-11 | 2.42E-08 |
|---|
| histone deacetylation | GO:0016575 |  | 6.943E-10 | 8.352E-07 |
|---|
| covalent chromatin modification | GO:0016569 |  | 9.878E-10 | 7.922E-07 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 2.457E-09 | 1.478E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.441E-08 | 3.58E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.144E-08 | 3.266E-05 |
|---|
| histone modification | GO:0016570 |  | 1.147E-07 | 3.941E-05 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 1.458E-06 | 4.386E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.542E-06 | 4.122E-04 |
|---|
| interphase | GO:0051325 |  | 1.746E-06 | 4.202E-04 |
|---|
| keratinocyte differentiation | GO:0030216 |  | 6.695E-06 | 1.464E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.887E-12 | 6.738E-09 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.198E-10 | 2.139E-07 |
|---|
| chromatin remodeling complex | GO:0016585 |  | 1.087E-09 | 1.294E-06 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 1.652E-09 | 1.475E-06 |
|---|
| kinase regulator activity | GO:0019207 |  | 4.256E-09 | 3.04E-06 |
|---|
| histone deacetylase complex | GO:0000118 |  | 5.387E-09 | 3.206E-06 |
|---|
| NuRD complex | GO:0016581 |  | 1.072E-08 | 5.466E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 2.997E-08 | 1.338E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.41E-08 | 1.75E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.823E-08 | 2.079E-05 |
|---|
| covalent chromatin modification | GO:0016569 |  | 5.823E-08 | 1.89E-05 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.911E-11 | 1.427E-07 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 2.062E-09 | 3.76E-06 |
|---|
| chromatin remodeling complex | GO:0016585 |  | 3.665E-09 | 4.457E-06 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 1.884E-08 | 1.718E-05 |
|---|
| histone deacetylase complex | GO:0000118 |  | 3.048E-08 | 2.224E-05 |
|---|
| kinase regulator activity | GO:0019207 |  | 3.862E-08 | 2.348E-05 |
|---|
| NuRD complex | GO:0016581 |  | 6.458E-08 | 3.366E-05 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.032E-07 | 4.708E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.038E-07 | 8.261E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.479E-07 | 9.045E-05 |
|---|
| covalent chromatin modification | GO:0016569 |  | 2.988E-07 | 9.911E-05 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.006E-11 | 2.42E-08 |
|---|
| histone deacetylation | GO:0016575 |  | 6.943E-10 | 8.352E-07 |
|---|
| covalent chromatin modification | GO:0016569 |  | 9.878E-10 | 7.922E-07 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 2.457E-09 | 1.478E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.441E-08 | 3.58E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.144E-08 | 3.266E-05 |
|---|
| histone modification | GO:0016570 |  | 1.147E-07 | 3.941E-05 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 1.458E-06 | 4.386E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.542E-06 | 4.122E-04 |
|---|
| interphase | GO:0051325 |  | 1.746E-06 | 4.202E-04 |
|---|
| keratinocyte differentiation | GO:0030216 |  | 6.695E-06 | 1.464E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.72E-11 | 1.776E-07 |
|---|
| histone deacetylation | GO:0016575 |  | 1.066E-09 | 1.226E-06 |
|---|
| covalent chromatin modification | GO:0016569 |  | 5.757E-09 | 4.414E-06 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 5.85E-09 | 3.364E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.87E-07 | 1.32E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.865E-07 | 1.482E-04 |
|---|
| histone modification | GO:0016570 |  | 5.096E-07 | 1.674E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 4.489E-06 | 1.291E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.381E-06 | 1.631E-03 |
|---|
| interphase | GO:0051325 |  | 6.994E-06 | 1.609E-03 |
|---|
| keratinocyte differentiation | GO:0030216 |  | 1.64E-05 | 3.43E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.72E-11 | 1.776E-07 |
|---|
| histone deacetylation | GO:0016575 |  | 1.066E-09 | 1.226E-06 |
|---|
| covalent chromatin modification | GO:0016569 |  | 5.757E-09 | 4.414E-06 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 5.85E-09 | 3.364E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.87E-07 | 1.32E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.865E-07 | 1.482E-04 |
|---|
| histone modification | GO:0016570 |  | 5.096E-07 | 1.674E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 4.489E-06 | 1.291E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.381E-06 | 1.631E-03 |
|---|
| interphase | GO:0051325 |  | 6.994E-06 | 1.609E-03 |
|---|
| keratinocyte differentiation | GO:0030216 |  | 1.64E-05 | 3.43E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.72E-11 | 1.776E-07 |
|---|
| histone deacetylation | GO:0016575 |  | 1.066E-09 | 1.226E-06 |
|---|
| covalent chromatin modification | GO:0016569 |  | 5.757E-09 | 4.414E-06 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 5.85E-09 | 3.364E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.87E-07 | 1.32E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.865E-07 | 1.482E-04 |
|---|
| histone modification | GO:0016570 |  | 5.096E-07 | 1.674E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 4.489E-06 | 1.291E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.381E-06 | 1.631E-03 |
|---|
| interphase | GO:0051325 |  | 6.994E-06 | 1.609E-03 |
|---|
| keratinocyte differentiation | GO:0030216 |  | 1.64E-05 | 3.43E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.72E-11 | 1.776E-07 |
|---|
| histone deacetylation | GO:0016575 |  | 1.066E-09 | 1.226E-06 |
|---|
| covalent chromatin modification | GO:0016569 |  | 5.757E-09 | 4.414E-06 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 5.85E-09 | 3.364E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.87E-07 | 1.32E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.865E-07 | 1.482E-04 |
|---|
| histone modification | GO:0016570 |  | 5.096E-07 | 1.674E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 4.489E-06 | 1.291E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.381E-06 | 1.631E-03 |
|---|
| interphase | GO:0051325 |  | 6.994E-06 | 1.609E-03 |
|---|
| keratinocyte differentiation | GO:0030216 |  | 1.64E-05 | 3.43E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.212E-10 | 2.876E-07 |
|---|
| histone deacetylation | GO:0016575 |  | 1.79E-09 | 2.123E-06 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 8.562E-09 | 6.772E-06 |
|---|
| covalent chromatin modification | GO:0016569 |  | 1.136E-08 | 6.739E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.926E-07 | 1.863E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.752E-07 | 1.88E-04 |
|---|
| histone modification | GO:0016570 |  | 8.014E-07 | 2.717E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 5.527E-06 | 1.639E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.463E-06 | 1.968E-03 |
|---|
| interphase | GO:0051325 |  | 8.169E-06 | 1.939E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 2.047E-05 | 4.417E-03 |
|---|



