Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 8721-10-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1897 | 5.837e-03 | 6.948e-03 | 6.577e-02 |
|---|
| IPC-NIBC-129 | 0.2556 | 7.540e-02 | 9.819e-02 | 5.814e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2743 | 1.703e-02 | 2.485e-02 | 3.571e-01 |
|---|
| Loi_GPL570 | 0.2179 | 5.693e-02 | 5.653e-02 | 2.802e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1485 | 6.000e-06 | 4.000e-05 | 3.300e-05 |
|---|
| Parker_GPL1390 | 0.2282 | 1.134e-01 | 1.142e-01 | 5.739e-01 |
|---|
| Parker_GPL887 | 0.0947 | 6.686e-01 | 6.958e-01 | 7.222e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3189 | 8.723e-02 | 6.241e-02 | 4.763e-01 |
|---|
| Schmidt | 0.2470 | 1.050e-02 | 1.496e-02 | 1.357e-01 |
|---|
| Sotiriou | 0.3122 | 5.089e-03 | 1.773e-03 | 1.030e-01 |
|---|
| Wang | 0.2117 | 1.332e-01 | 9.609e-02 | 4.387e-01 |
|---|
| Zhang | 0.2844 | 6.625e-02 | 4.000e-02 | 1.655e-01 |
|---|
| Zhou | 0.3699 | 9.043e-02 | 9.903e-02 | 5.140e-01 |
|---|
Expression data for subnetwork 8721-10-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
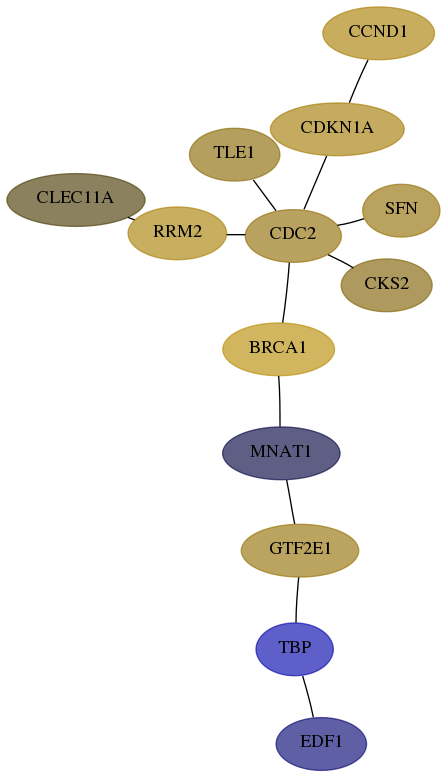
Score for each gene in subnetwork 8721-10-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| tbp |   | 2 | 104 | 322 | 308 | -0.139 | 0.211 | 0.041 | 0.154 | 0.096 | 0.049 | -0.384 | -0.102 | 0.080 | -0.033 | -0.024 | 0.061 | 0.160 |
|---|
| rrm2 |   | 21 | 10 | 44 | 26 | 0.131 | 0.207 | 0.227 | 0.187 | 0.161 | 0.178 | 0.137 | 0.303 | 0.190 | 0.219 | 0.117 | 0.129 | 0.302 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| tle1 |   | 23 | 9 | 78 | 60 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | -0.028 | 0.068 | 0.025 |
|---|
| brca1 |   | 8 | 33 | 175 | 150 | 0.192 | 0.052 | 0.112 | 0.022 | 0.125 | -0.124 | 0.305 | 0.113 | 0.077 | 0.168 | 0.155 | 0.064 | 0.209 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| cks2 |   | 44 | 6 | 73 | 54 | 0.050 | 0.233 | 0.170 | 0.103 | 0.186 | 0.172 | 0.093 | 0.167 | 0.263 | 0.151 | 0.145 | 0.284 | 0.383 |
|---|
| gtf2e1 |   | 2 | 104 | 322 | 308 | 0.080 | 0.101 | 0.138 | 0.034 | 0.106 | -0.011 | -0.129 | 0.182 | 0.073 | 0.183 | 0.060 | 0.048 | 0.085 |
|---|
| edf1 |   | 1 | 164 | 322 | 328 | -0.036 | 0.087 | 0.112 | 0.146 | 0.072 | -0.019 | -0.109 | 0.131 | 0.068 | 0.007 | 0.065 | 0.093 | 0.090 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| sfn |   | 50 | 4 | 29 | 17 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.025 | 0.079 | 0.181 |
|---|
| clec11a |   | 17 | 12 | 44 | 27 | 0.013 | 0.101 | 0.071 | -0.085 | 0.066 | 0.082 | 0.209 | -0.126 | -0.034 | -0.001 | 0.191 | 0.059 | -0.075 |
|---|
| mnat1 |   | 3 | 79 | 175 | 162 | -0.011 | 0.083 | 0.075 | 0.305 | 0.105 | 0.111 | -0.009 | 0.107 | 0.118 | -0.002 | 0.144 | 0.307 | 0.086 |
|---|
GO Enrichment output for subnetwork 8721-10-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.881E-11 | 1.353E-07 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.534E-08 | 5.215E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 7.928E-08 | 6.078E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.415E-07 | 1.389E-04 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 4.683E-07 | 2.154E-04 |
|---|
| RNA elongation | GO:0006354 |  | 5.553E-07 | 2.129E-04 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 2.357E-06 | 7.746E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.975E-06 | 8.553E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 3.954E-06 | 1.011E-03 |
|---|
| transcription initiation | GO:0006352 |  | 5.135E-06 | 1.181E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.802E-05 | 3.767E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.912E-12 | 7.113E-09 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.283E-08 | 4.01E-05 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 5.944E-08 | 4.84E-05 |
|---|
| RNA elongation | GO:0006354 |  | 8.048E-08 | 4.915E-05 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 3.601E-07 | 1.759E-04 |
|---|
| transcription initiation | GO:0006352 |  | 7.439E-07 | 3.029E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 7.906E-07 | 2.759E-04 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 5.138E-06 | 1.569E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.165E-06 | 1.402E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 6.26E-06 | 1.529E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 7.704E-06 | 1.711E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.834E-12 | 1.885E-08 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.994E-08 | 2.399E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.781E-08 | 3.834E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.345E-08 | 3.817E-05 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 1.061E-07 | 5.103E-05 |
|---|
| RNA elongation | GO:0006354 |  | 1.444E-07 | 5.79E-05 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 5.959E-07 | 2.048E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 8.7E-07 | 2.617E-04 |
|---|
| transcription initiation | GO:0006352 |  | 1.262E-06 | 3.374E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 1.297E-06 | 3.121E-04 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 7.155E-06 | 1.565E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.912E-12 | 7.113E-09 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.283E-08 | 4.01E-05 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 5.944E-08 | 4.84E-05 |
|---|
| RNA elongation | GO:0006354 |  | 8.048E-08 | 4.915E-05 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 3.601E-07 | 1.759E-04 |
|---|
| transcription initiation | GO:0006352 |  | 7.439E-07 | 3.029E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 7.906E-07 | 2.759E-04 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 5.138E-06 | 1.569E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.165E-06 | 1.402E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 6.26E-06 | 1.529E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 7.704E-06 | 1.711E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.834E-12 | 1.885E-08 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.994E-08 | 2.399E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.781E-08 | 3.834E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.345E-08 | 3.817E-05 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 1.061E-07 | 5.103E-05 |
|---|
| RNA elongation | GO:0006354 |  | 1.444E-07 | 5.79E-05 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 5.959E-07 | 2.048E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 8.7E-07 | 2.617E-04 |
|---|
| transcription initiation | GO:0006352 |  | 1.262E-06 | 3.374E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 1.297E-06 | 3.121E-04 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 7.155E-06 | 1.565E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.887E-12 | 6.738E-09 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.198E-10 | 2.139E-07 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 1.652E-09 | 1.967E-06 |
|---|
| kinase regulator activity | GO:0019207 |  | 4.256E-09 | 3.8E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 2.997E-08 | 2.141E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.41E-08 | 2.625E-05 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 5.823E-08 | 2.971E-05 |
|---|
| RNA elongation | GO:0006354 |  | 7.55E-08 | 3.37E-05 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 1.942E-07 | 7.707E-05 |
|---|
| DNA-directed RNA polymerase II. holoenzyme | GO:0016591 |  | 2.108E-07 | 7.528E-05 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 2.707E-07 | 8.789E-05 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.939E-11 | 1.072E-07 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.719E-09 | 3.135E-06 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 1.496E-08 | 1.819E-05 |
|---|
| kinase regulator activity | GO:0019207 |  | 3.067E-08 | 2.797E-05 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 9.036E-08 | 6.593E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.701E-07 | 1.034E-04 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 2.275E-07 | 1.185E-04 |
|---|
| RNA elongation | GO:0006354 |  | 2.982E-07 | 1.36E-04 |
|---|
| DNA-directed RNA polymerase II. holoenzyme | GO:0016591 |  | 7.534E-07 | 3.054E-04 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 8.625E-07 | 3.146E-04 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 8.978E-07 | 2.977E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.834E-12 | 1.885E-08 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.994E-08 | 2.399E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.781E-08 | 3.834E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.345E-08 | 3.817E-05 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 1.061E-07 | 5.103E-05 |
|---|
| RNA elongation | GO:0006354 |  | 1.444E-07 | 5.79E-05 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 5.959E-07 | 2.048E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 8.7E-07 | 2.617E-04 |
|---|
| transcription initiation | GO:0006352 |  | 1.262E-06 | 3.374E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 1.297E-06 | 3.121E-04 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 7.155E-06 | 1.565E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.881E-11 | 1.353E-07 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.534E-08 | 5.215E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 7.928E-08 | 6.078E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.415E-07 | 1.389E-04 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 4.683E-07 | 2.154E-04 |
|---|
| RNA elongation | GO:0006354 |  | 5.553E-07 | 2.129E-04 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 2.357E-06 | 7.746E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.975E-06 | 8.553E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 3.954E-06 | 1.011E-03 |
|---|
| transcription initiation | GO:0006352 |  | 5.135E-06 | 1.181E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.802E-05 | 3.767E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.881E-11 | 1.353E-07 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.534E-08 | 5.215E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 7.928E-08 | 6.078E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.415E-07 | 1.389E-04 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 4.683E-07 | 2.154E-04 |
|---|
| RNA elongation | GO:0006354 |  | 5.553E-07 | 2.129E-04 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 2.357E-06 | 7.746E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.975E-06 | 8.553E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 3.954E-06 | 1.011E-03 |
|---|
| transcription initiation | GO:0006352 |  | 5.135E-06 | 1.181E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.802E-05 | 3.767E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.881E-11 | 1.353E-07 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.534E-08 | 5.215E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 7.928E-08 | 6.078E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.415E-07 | 1.389E-04 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 4.683E-07 | 2.154E-04 |
|---|
| RNA elongation | GO:0006354 |  | 5.553E-07 | 2.129E-04 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 2.357E-06 | 7.746E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.975E-06 | 8.553E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 3.954E-06 | 1.011E-03 |
|---|
| transcription initiation | GO:0006352 |  | 5.135E-06 | 1.181E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.802E-05 | 3.767E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.881E-11 | 1.353E-07 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.534E-08 | 5.215E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 7.928E-08 | 6.078E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.415E-07 | 1.389E-04 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 4.683E-07 | 2.154E-04 |
|---|
| RNA elongation | GO:0006354 |  | 5.553E-07 | 2.129E-04 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 2.357E-06 | 7.746E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.975E-06 | 8.553E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 3.954E-06 | 1.011E-03 |
|---|
| transcription initiation | GO:0006352 |  | 5.135E-06 | 1.181E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.802E-05 | 3.767E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.91E-11 | 1.402E-07 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 6.918E-08 | 8.208E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.106E-07 | 8.748E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.483E-07 | 1.473E-04 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 4.671E-07 | 2.217E-04 |
|---|
| RNA elongation | GO:0006354 |  | 5.499E-07 | 2.175E-04 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 2.481E-06 | 8.411E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.005E-06 | 8.912E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 3.942E-06 | 1.039E-03 |
|---|
| transcription initiation | GO:0006352 |  | 5.432E-06 | 1.289E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 1.639E-05 | 3.537E-03 |
|---|



