Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 79577-9-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2600 | 5.826e-03 | 6.934e-03 | 6.567e-02 |
|---|
| IPC-NIBC-129 | 0.2726 | 7.140e-02 | 9.274e-02 | 5.638e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2744 | 2.304e-02 | 3.262e-02 | 4.189e-01 |
|---|
| Loi_GPL570 | 0.2036 | 4.509e-02 | 4.474e-02 | 2.360e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1543 | 2.020e-04 | 9.170e-04 | 5.447e-03 |
|---|
| Parker_GPL1390 | 0.2322 | 4.209e-02 | 3.864e-02 | 3.420e-01 |
|---|
| Parker_GPL887 | 0.1076 | 6.222e-01 | 6.517e-01 | 6.722e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3487 | 2.425e-01 | 2.039e-01 | 8.048e-01 |
|---|
| Schmidt | 0.1953 | 7.451e-03 | 1.078e-02 | 1.038e-01 |
|---|
| Sotiriou | 0.3331 | 9.130e-04 | 2.020e-04 | 2.952e-02 |
|---|
| Wang | 0.1822 | 1.214e-01 | 8.622e-02 | 4.139e-01 |
|---|
| Zhang | 0.1828 | 1.014e-01 | 6.662e-02 | 2.434e-01 |
|---|
| Zhou | 0.4732 | 6.831e-02 | 7.298e-02 | 4.146e-01 |
|---|
Expression data for subnetwork 79577-9-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
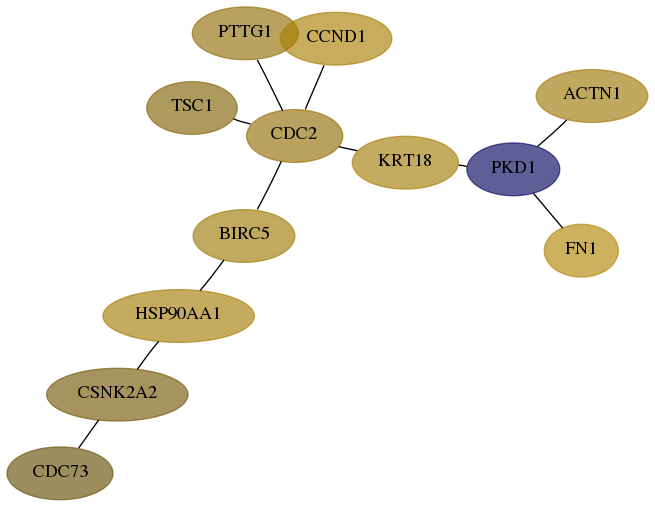
Score for each gene in subnetwork 79577-9-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| cdc73 |   | 1 | 164 | 234 | 240 | 0.027 | 0.222 | 0.127 | -0.186 | 0.079 | 0.011 | -0.048 | 0.164 | 0.111 | 0.096 | 0.175 | 0.011 | 0.115 |
|---|
| pttg1 |   | 10 | 25 | 31 | 21 | 0.072 | 0.270 | 0.175 | 0.183 | 0.200 | 0.208 | 0.347 | 0.334 | 0.224 | 0.204 | 0.147 | 0.210 | 0.309 |
|---|
| csnk2a2 |   | 1 | 164 | 234 | 240 | 0.036 | 0.182 | 0.089 | -0.033 | 0.074 | 0.094 | -0.125 | 0.149 | 0.060 | 0.090 | 0.060 | -0.155 | 0.174 |
|---|
| krt18 |   | 15 | 15 | 230 | 195 | 0.112 | -0.078 | 0.147 | 0.150 | 0.161 | -0.086 | -0.204 | 0.083 | -0.099 | 0.118 | 0.086 | -0.114 | 0.222 |
|---|
| hsp90aa1 |   | 11 | 21 | 234 | 204 | 0.116 | 0.030 | 0.164 | 0.076 | 0.119 | 0.155 | -0.199 | 0.169 | 0.069 | 0.102 | 0.052 | -0.011 | 0.399 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| tsc1 |   | 47 | 5 | 17 | 8 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.043 | -0.007 | 0.207 |
|---|
| pkd1 |   | 9 | 27 | 234 | 205 | -0.021 | 0.055 | 0.007 | 0.037 | 0.071 | 0.184 | 0.256 | -0.217 | -0.068 | -0.001 | 0.046 | 0.077 | 0.151 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| actn1 |   | 4 | 60 | 1 | 9 | 0.100 | 0.223 | 0.031 | -0.052 | 0.053 | 0.156 | 0.031 | 0.019 | 0.024 | 0.027 | 0.124 | -0.008 | 0.241 |
|---|
| fn1 |   | 3 | 79 | 234 | 222 | 0.156 | 0.240 | -0.023 | -0.035 | 0.017 | 0.089 | -0.120 | 0.077 | -0.045 | -0.063 | 0.109 | 0.067 | 0.230 |
|---|
| birc5 |   | 14 | 17 | 162 | 140 | 0.101 | 0.216 | 0.190 | 0.134 | 0.223 | 0.149 | 0.166 | 0.202 | 0.236 | 0.241 | 0.124 | 0.205 | 0.400 |
|---|
GO Enrichment output for subnetwork 79577-9-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.121E-05 | 0.0487855 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 2.121E-05 | 0.02439275 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 3.179E-05 | 0.02437043 |
|---|
| protein refolding | GO:0042026 |  | 3.179E-05 | 0.01827782 |
|---|
| spindle checkpoint | GO:0031577 |  | 3.179E-05 | 0.01462226 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.976E-05 | 0.0152399 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.446E-05 | 0.01460888 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 4.446E-05 | 0.01278277 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 4.446E-05 | 0.01136246 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 4.446E-05 | 0.01022621 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 5.923E-05 | 0.01238407 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.876E-06 | 0.01679724 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 9.146E-06 | 0.01117171 |
|---|
| protein refolding | GO:0042026 |  | 1.031E-05 | 8.394E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 1.031E-05 | 6.296E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 1.442E-05 | 7.047E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 1.442E-05 | 5.873E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.442E-05 | 5.034E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.442E-05 | 4.405E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.922E-05 | 5.218E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.922E-05 | 4.696E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.922E-05 | 4.269E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.937E-06 | 0.02150125 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 8.937E-06 | 0.01075063 |
|---|
| protein refolding | GO:0042026 |  | 1.34E-05 | 0.0107442 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.348E-05 | 8.111E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 1.874E-05 | 9.02E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 1.874E-05 | 7.516E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.874E-05 | 6.443E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.874E-05 | 5.637E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.498E-05 | 6.677E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 2.498E-05 | 6.01E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 2.498E-05 | 5.463E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.876E-06 | 0.01679724 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 9.146E-06 | 0.01117171 |
|---|
| protein refolding | GO:0042026 |  | 1.031E-05 | 8.394E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 1.031E-05 | 6.296E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 1.442E-05 | 7.047E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 1.442E-05 | 5.873E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.442E-05 | 5.034E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.442E-05 | 4.405E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.922E-05 | 5.218E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.922E-05 | 4.696E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.922E-05 | 4.269E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.937E-06 | 0.02150125 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 8.937E-06 | 0.01075063 |
|---|
| protein refolding | GO:0042026 |  | 1.34E-05 | 0.0107442 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.348E-05 | 8.111E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 1.874E-05 | 9.02E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 1.874E-05 | 7.516E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.874E-05 | 6.443E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.874E-05 | 5.637E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.498E-05 | 6.677E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 2.498E-05 | 6.01E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 2.498E-05 | 5.463E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of Ras GTPase activity | GO:0032318 |  | 2.583E-07 | 9.224E-04 |
|---|
| regulation of GTPase activity | GO:0043087 |  | 5.622E-07 | 1.004E-03 |
|---|
| cysteine-type endopeptidase inhibitor activity | GO:0004869 |  | 1.243E-06 | 1.48E-03 |
|---|
| regulation of Rab protein signal transduction | GO:0032483 |  | 3.013E-06 | 2.69E-03 |
|---|
| Rab GTPase activator activity | GO:0005097 |  | 3.249E-06 | 2.321E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 5.061E-06 | 3.012E-03 |
|---|
| nitric-oxide synthase regulator activity | GO:0030235 |  | 5.061E-06 | 2.582E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.061E-06 | 2.259E-03 |
|---|
| protein N-terminus binding | GO:0047485 |  | 5.257E-06 | 2.086E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.589E-06 | 2.71E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 7.589E-06 | 2.464E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| midbody | GO:0030496 |  | 5.735E-08 | 2.092E-04 |
|---|
| spindle microtubule | GO:0005876 |  | 2.122E-07 | 3.871E-04 |
|---|
| chaperone binding | GO:0051087 |  | 2.962E-07 | 3.602E-04 |
|---|
| microtubule organizing center part | GO:0044450 |  | 1.283E-06 | 1.17E-03 |
|---|
| cysteine-type endopeptidase inhibitor activity | GO:0004869 |  | 1.41E-06 | 1.029E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.177E-06 | 1.932E-03 |
|---|
| protein N-terminus binding | GO:0047485 |  | 4.444E-06 | 2.316E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 4.447E-06 | 2.028E-03 |
|---|
| nitric-oxide synthase regulator activity | GO:0030235 |  | 4.447E-06 | 1.802E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 4.447E-06 | 1.622E-03 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 4.447E-06 | 1.475E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.937E-06 | 0.02150125 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 8.937E-06 | 0.01075063 |
|---|
| protein refolding | GO:0042026 |  | 1.34E-05 | 0.0107442 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.348E-05 | 8.111E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 1.874E-05 | 9.02E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 1.874E-05 | 7.516E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.874E-05 | 6.443E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.874E-05 | 5.637E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.498E-05 | 6.677E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 2.498E-05 | 6.01E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 2.498E-05 | 5.463E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.121E-05 | 0.0487855 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 2.121E-05 | 0.02439275 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 3.179E-05 | 0.02437043 |
|---|
| protein refolding | GO:0042026 |  | 3.179E-05 | 0.01827782 |
|---|
| spindle checkpoint | GO:0031577 |  | 3.179E-05 | 0.01462226 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.976E-05 | 0.0152399 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.446E-05 | 0.01460888 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 4.446E-05 | 0.01278277 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 4.446E-05 | 0.01136246 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 4.446E-05 | 0.01022621 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 5.923E-05 | 0.01238407 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.121E-05 | 0.0487855 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 2.121E-05 | 0.02439275 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 3.179E-05 | 0.02437043 |
|---|
| protein refolding | GO:0042026 |  | 3.179E-05 | 0.01827782 |
|---|
| spindle checkpoint | GO:0031577 |  | 3.179E-05 | 0.01462226 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.976E-05 | 0.0152399 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.446E-05 | 0.01460888 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 4.446E-05 | 0.01278277 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 4.446E-05 | 0.01136246 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 4.446E-05 | 0.01022621 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 5.923E-05 | 0.01238407 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.121E-05 | 0.0487855 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 2.121E-05 | 0.02439275 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 3.179E-05 | 0.02437043 |
|---|
| protein refolding | GO:0042026 |  | 3.179E-05 | 0.01827782 |
|---|
| spindle checkpoint | GO:0031577 |  | 3.179E-05 | 0.01462226 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.976E-05 | 0.0152399 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.446E-05 | 0.01460888 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 4.446E-05 | 0.01278277 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 4.446E-05 | 0.01136246 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 4.446E-05 | 0.01022621 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 5.923E-05 | 0.01238407 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.121E-05 | 0.0487855 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 2.121E-05 | 0.02439275 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 3.179E-05 | 0.02437043 |
|---|
| protein refolding | GO:0042026 |  | 3.179E-05 | 0.01827782 |
|---|
| spindle checkpoint | GO:0031577 |  | 3.179E-05 | 0.01462226 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.976E-05 | 0.0152399 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.446E-05 | 0.01460888 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 4.446E-05 | 0.01278277 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 4.446E-05 | 0.01136246 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 4.446E-05 | 0.01022621 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 5.923E-05 | 0.01238407 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| detection of mechanical stimulus | GO:0050982 |  | 3.419E-09 | 8.113E-06 |
|---|
| cartilage condensation | GO:0001502 |  | 4.781E-09 | 5.673E-06 |
|---|
| protein refolding | GO:0042026 |  | 6.836E-08 | 5.408E-05 |
|---|
| cell-matrix adhesion | GO:0007160 |  | 7.943E-08 | 4.712E-05 |
|---|
| cell-substrate adhesion | GO:0031589 |  | 1.121E-07 | 5.32E-05 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.195E-07 | 4.727E-05 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.91E-07 | 6.475E-05 |
|---|
| response to mechanical stimulus | GO:0009612 |  | 2.167E-07 | 6.429E-05 |
|---|
| JAK-STAT cascade | GO:0007259 |  | 6.276E-07 | 1.655E-04 |
|---|
| detection of abiotic stimulus | GO:0009582 |  | 8.956E-07 | 2.125E-04 |
|---|
| cartilage development | GO:0051216 |  | 1.676E-06 | 3.616E-04 |
|---|



