Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 7818-9-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1752 | 5.218e-03 | 6.205e-03 | 6.029e-02 |
|---|
| IPC-NIBC-129 | 0.2518 | 1.983e-02 | 2.355e-02 | 2.242e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2797 | 4.044e-03 | 6.820e-03 | 1.480e-01 |
|---|
| Loi_GPL570 | 0.2852 | 5.999e-02 | 5.957e-02 | 2.910e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1606 | 7.000e-06 | 4.700e-05 | 4.300e-05 |
|---|
| Parker_GPL1390 | 0.3247 | 1.385e-01 | 1.418e-01 | 6.281e-01 |
|---|
| Parker_GPL887 | 0.1674 | 4.365e-01 | 4.712e-01 | 4.623e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3320 | 1.938e-01 | 1.574e-01 | 7.345e-01 |
|---|
| Schmidt | 0.2444 | 1.430e-02 | 2.011e-02 | 1.715e-01 |
|---|
| Sotiriou | 0.2933 | 1.052e-02 | 4.421e-03 | 1.678e-01 |
|---|
| Wang | 0.1758 | 1.096e-01 | 7.654e-02 | 3.878e-01 |
|---|
| Zhang | 0.2057 | 1.112e-01 | 7.439e-02 | 2.639e-01 |
|---|
| Zhou | 0.3257 | 7.998e-02 | 8.667e-02 | 4.693e-01 |
|---|
Expression data for subnetwork 7818-9-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
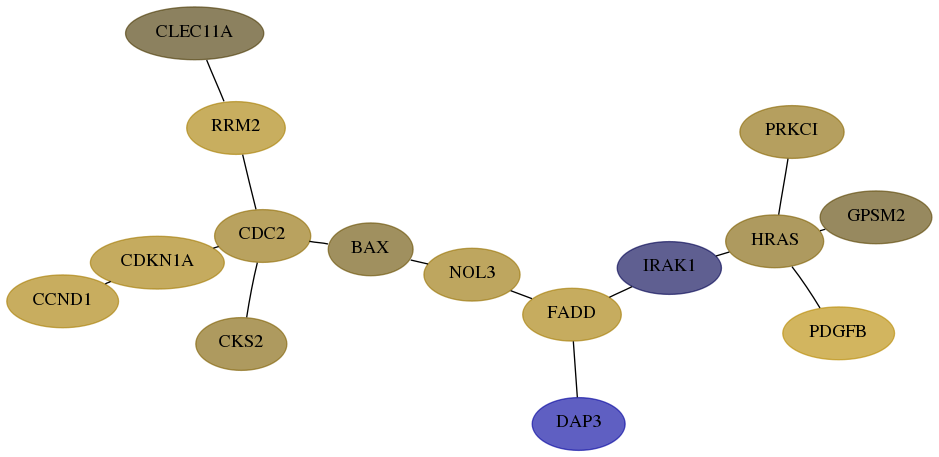
Score for each gene in subnetwork 7818-9-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| pdgfb |   | 2 | 104 | 161 | 156 | 0.186 | -0.069 | 0.089 | 0.269 | 0.087 | 0.107 | -0.031 | 0.163 | 0.038 | 0.139 | -0.163 | 0.037 | 0.170 |
|---|
| irak1 |   | 3 | 79 | 120 | 112 | -0.016 | 0.242 | 0.007 | 0.133 | 0.108 | 0.303 | 0.257 | 0.133 | 0.177 | 0.084 | -0.046 | -0.026 | 0.081 |
|---|
| rrm2 |   | 21 | 10 | 44 | 26 | 0.131 | 0.207 | 0.227 | 0.187 | 0.161 | 0.178 | 0.137 | 0.303 | 0.190 | 0.219 | 0.117 | 0.129 | 0.302 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| bax |   | 9 | 27 | 17 | 15 | 0.029 | 0.185 | 0.233 | 0.192 | 0.088 | -0.043 | 0.184 | 0.172 | 0.175 | 0.019 | -0.094 | 0.041 | 0.235 |
|---|
| prkci |   | 12 | 20 | 31 | 19 | 0.065 | 0.104 | 0.012 | 0.170 | 0.112 | 0.088 | -0.103 | -0.170 | 0.196 | 0.128 | -0.054 | 0.124 | 0.149 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| fadd |   | 5 | 50 | 64 | 58 | 0.121 | -0.009 | 0.155 | 0.307 | 0.166 | 0.119 | 0.084 | 0.300 | 0.070 | 0.116 | 0.100 | 0.032 | 0.243 |
|---|
| cks2 |   | 44 | 6 | 73 | 54 | 0.050 | 0.233 | 0.170 | 0.103 | 0.186 | 0.172 | 0.093 | 0.167 | 0.263 | 0.151 | 0.145 | 0.284 | 0.383 |
|---|
| dap3 |   | 1 | 164 | 228 | 236 | -0.105 | 0.187 | -0.036 | -0.058 | 0.070 | -0.011 | 0.197 | 0.211 | -0.015 | 0.010 | 0.108 | 0.002 | 0.159 |
|---|
| gpsm2 |   | 4 | 60 | 120 | 108 | 0.021 | 0.286 | 0.155 | 0.144 | 0.108 | 0.242 | 0.307 | 0.134 | 0.234 | 0.077 | 0.176 | 0.098 | 0.207 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| hras |   | 7 | 38 | 88 | 75 | 0.049 | -0.030 | 0.094 | 0.210 | 0.160 | 0.195 | 0.022 | 0.245 | 0.086 | 0.208 | 0.001 | 0.003 | 0.197 |
|---|
| nol3 |   | 3 | 79 | 228 | 221 | 0.089 | 0.110 | 0.102 | -0.119 | 0.094 | 0.021 | 0.040 | 0.125 | -0.023 | 0.016 | 0.180 | 0.059 | 0.069 |
|---|
| clec11a |   | 17 | 12 | 44 | 27 | 0.013 | 0.101 | 0.071 | -0.085 | 0.066 | 0.082 | 0.209 | -0.126 | -0.034 | -0.001 | 0.191 | 0.059 | -0.075 |
|---|
GO Enrichment output for subnetwork 7818-9-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 6.55E-08 | 1.507E-04 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.145E-07 | 1.317E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.073E-06 | 8.225E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 2.864E-06 | 1.647E-03 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 3.131E-06 | 1.44E-03 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 4.289E-06 | 1.644E-03 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 4.961E-06 | 1.63E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 1.36E-05 | 3.91E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 2.291E-05 | 5.854E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 3.433E-05 | 7.895E-03 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 3.433E-05 | 7.177E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mitochondrion localization | GO:0051646 |  | 6.101E-08 | 1.49E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.348E-07 | 2.868E-04 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 1.654E-06 | 1.347E-03 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 2.351E-06 | 1.436E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 8.344E-06 | 4.077E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.077E-05 | 4.385E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 1.251E-05 | 4.366E-03 |
|---|
| negative regulation of platelet activation | GO:0010544 |  | 1.251E-05 | 3.82E-03 |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 1.387E-05 | 3.765E-03 |
|---|
| regulation of neuron apoptosis | GO:0043523 |  | 1.655E-05 | 4.042E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.75E-05 | 3.887E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.113E-08 | 7.49E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 7.461E-08 | 8.976E-05 |
|---|
| mitochondrion localization | GO:0051646 |  | 7.461E-08 | 5.984E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.045E-07 | 1.832E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 1.084E-06 | 5.215E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.356E-06 | 5.438E-04 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 1.78E-06 | 6.117E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 2.567E-06 | 7.72E-04 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 9.574E-06 | 2.56E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.315E-05 | 3.164E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 1.435E-05 | 3.139E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mitochondrion localization | GO:0051646 |  | 6.101E-08 | 1.49E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.348E-07 | 2.868E-04 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 1.654E-06 | 1.347E-03 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 2.351E-06 | 1.436E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 8.344E-06 | 4.077E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.077E-05 | 4.385E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 1.251E-05 | 4.366E-03 |
|---|
| negative regulation of platelet activation | GO:0010544 |  | 1.251E-05 | 3.82E-03 |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 1.387E-05 | 3.765E-03 |
|---|
| regulation of neuron apoptosis | GO:0043523 |  | 1.655E-05 | 4.042E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.75E-05 | 3.887E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.113E-08 | 7.49E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 7.461E-08 | 8.976E-05 |
|---|
| mitochondrion localization | GO:0051646 |  | 7.461E-08 | 5.984E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.045E-07 | 1.832E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 1.084E-06 | 5.215E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.356E-06 | 5.438E-04 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 1.78E-06 | 6.117E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 2.567E-06 | 7.72E-04 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 9.574E-06 | 2.56E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.315E-05 | 3.164E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 1.435E-05 | 3.139E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.676E-10 | 5.987E-07 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 3.814E-08 | 6.809E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.145E-07 | 1.363E-04 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 3.136E-07 | 2.8E-04 |
|---|
| kinase regulator activity | GO:0019207 |  | 6.633E-07 | 4.737E-04 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 8.992E-07 | 5.352E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 1.366E-06 | 6.969E-04 |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 6.117E-06 | 2.73E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 8.371E-06 | 3.321E-03 |
|---|
| regulation of neuron apoptosis | GO:0043523 |  | 9.465E-06 | 3.38E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.012E-05 | 3.285E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 2.659E-10 | 9.699E-07 |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 2.159E-08 | 3.938E-05 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 2.325E-08 | 2.827E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.523E-08 | 5.949E-05 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 1.868E-07 | 1.363E-04 |
|---|
| kinase regulator activity | GO:0019207 |  | 3.309E-07 | 2.012E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 3.489E-07 | 1.818E-04 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 4.002E-07 | 1.825E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 5.488E-07 | 2.224E-04 |
|---|
| response to light stimulus | GO:0009416 |  | 1E-06 | 3.648E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 1.07E-06 | 3.549E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.113E-08 | 7.49E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 7.461E-08 | 8.976E-05 |
|---|
| mitochondrion localization | GO:0051646 |  | 7.461E-08 | 5.984E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.045E-07 | 1.832E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 1.084E-06 | 5.215E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.356E-06 | 5.438E-04 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 1.78E-06 | 6.117E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 2.567E-06 | 7.72E-04 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 9.574E-06 | 2.56E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.315E-05 | 3.164E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 1.435E-05 | 3.139E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 6.55E-08 | 1.507E-04 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.145E-07 | 1.317E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.073E-06 | 8.225E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 2.864E-06 | 1.647E-03 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 3.131E-06 | 1.44E-03 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 4.289E-06 | 1.644E-03 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 4.961E-06 | 1.63E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 1.36E-05 | 3.91E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 2.291E-05 | 5.854E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 3.433E-05 | 7.895E-03 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 3.433E-05 | 7.177E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 6.55E-08 | 1.507E-04 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.145E-07 | 1.317E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.073E-06 | 8.225E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 2.864E-06 | 1.647E-03 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 3.131E-06 | 1.44E-03 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 4.289E-06 | 1.644E-03 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 4.961E-06 | 1.63E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 1.36E-05 | 3.91E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 2.291E-05 | 5.854E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 3.433E-05 | 7.895E-03 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 3.433E-05 | 7.177E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 6.55E-08 | 1.507E-04 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.145E-07 | 1.317E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.073E-06 | 8.225E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 2.864E-06 | 1.647E-03 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 3.131E-06 | 1.44E-03 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 4.289E-06 | 1.644E-03 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 4.961E-06 | 1.63E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 1.36E-05 | 3.91E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 2.291E-05 | 5.854E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 3.433E-05 | 7.895E-03 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 3.433E-05 | 7.177E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 6.55E-08 | 1.507E-04 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.145E-07 | 1.317E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.073E-06 | 8.225E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 2.864E-06 | 1.647E-03 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 3.131E-06 | 1.44E-03 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 4.289E-06 | 1.644E-03 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 4.961E-06 | 1.63E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 1.36E-05 | 3.91E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 2.291E-05 | 5.854E-03 |
|---|
| photoreceptor cell development | GO:0042461 |  | 3.433E-05 | 7.895E-03 |
|---|
| regulation of Rac protein signal transduction | GO:0035020 |  | 3.433E-05 | 7.177E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 2.144E-08 | 5.089E-05 |
|---|
| negative regulation of platelet activation | GO:0010544 |  | 2.782E-08 | 3.301E-05 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 4.514E-08 | 3.571E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 9.718E-08 | 5.765E-05 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 9.718E-08 | 4.612E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.553E-07 | 6.144E-05 |
|---|
| negative regulation of lipid biosynthetic process | GO:0051055 |  | 2.328E-07 | 7.891E-05 |
|---|
| regulation of smooth muscle cell migration | GO:0014910 |  | 2.328E-07 | 6.905E-05 |
|---|
| positive regulation of endothelial cell migration | GO:0010595 |  | 3.322E-07 | 8.759E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 3.322E-07 | 7.883E-05 |
|---|
| positive regulation of endothelial cell proliferation | GO:0001938 |  | 6.079E-07 | 1.311E-04 |
|---|



