Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6513-8-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2174 | 8.209e-03 | 9.799e-03 | 8.539e-02 |
|---|
| IPC-NIBC-129 | 0.2837 | 8.876e-02 | 1.164e-01 | 6.346e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2579 | 1.138e-02 | 1.729e-02 | 2.842e-01 |
|---|
| Loi_GPL570 | 0.2369 | 3.870e-02 | 3.840e-02 | 2.103e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1720 | 7.600e-05 | 3.900e-04 | 1.479e-03 |
|---|
| Parker_GPL1390 | 0.2161 | 7.697e-02 | 7.495e-02 | 4.748e-01 |
|---|
| Parker_GPL887 | 0.1769 | 4.117e-01 | 4.465e-01 | 4.335e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3615 | 1.262e-01 | 9.576e-02 | 5.931e-01 |
|---|
| Schmidt | 0.2097 | 1.308e-02 | 1.847e-02 | 1.605e-01 |
|---|
| Sotiriou | 0.2988 | 1.450e-03 | 3.630e-04 | 4.180e-02 |
|---|
| Wang | 0.1038 | 9.128e-02 | 6.182e-02 | 3.443e-01 |
|---|
| Zhang | 0.2484 | 3.041e-01 | 2.474e-01 | 5.886e-01 |
|---|
| Zhou | 0.4455 | 6.044e-02 | 6.384e-02 | 3.745e-01 |
|---|
Expression data for subnetwork 6513-8-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
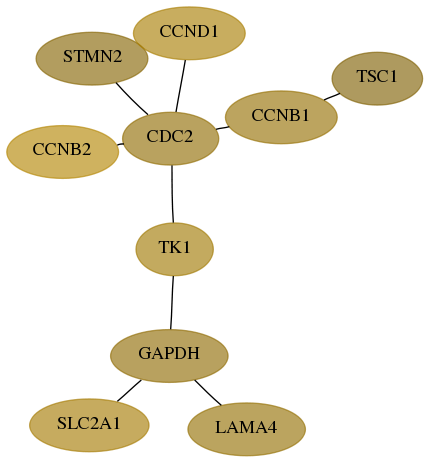
Score for each gene in subnetwork 6513-8-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| ccnb2 |   | 16 | 13 | 153 | 125 | 0.164 | 0.176 | 0.204 | 0.123 | 0.172 | 0.178 | 0.090 | 0.366 | 0.240 | 0.169 | 0.144 | 0.211 | 0.463 |
|---|
| stmn2 |   | 29 | 8 | 1 | 3 | 0.057 | 0.106 | 0.005 | 0.052 | 0.110 | 0.071 | -0.099 | 0.006 | 0.034 | 0.103 | 0.075 | 0.113 | -0.039 |
|---|
| ccnb1 |   | 36 | 7 | 78 | 59 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | 0.152 | 0.167 | 0.398 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| lama4 |   | 5 | 50 | 136 | 121 | 0.079 | 0.190 | 0.023 | -0.039 | 0.041 | 0.126 | -0.167 | -0.168 | -0.007 | -0.089 | 0.200 | 0.057 | 0.089 |
|---|
| tsc1 |   | 47 | 5 | 17 | 8 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.043 | -0.007 | 0.207 |
|---|
| gapdh |   | 6 | 43 | 136 | 117 | 0.071 | 0.312 | 0.102 | 0.145 | 0.165 | 0.172 | -0.253 | 0.288 | 0.064 | 0.213 | 0.056 | -0.088 | 0.235 |
|---|
| tk1 |   | 6 | 43 | 1 | 7 | 0.109 | 0.149 | 0.190 | -0.017 | 0.131 | 0.105 | -0.100 | 0.257 | 0.204 | 0.142 | 0.064 | 0.164 | 0.222 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| slc2a1 |   | 2 | 104 | 136 | 139 | 0.118 | 0.267 | 0.125 | 0.260 | 0.126 | 0.065 | -0.206 | 0.154 | 0.069 | 0.098 | 0.075 | -0.081 | 0.006 |
|---|
GO Enrichment output for subnetwork 6513-8-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.117E-05 | 0.02568926 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.117E-05 | 0.01284463 |
|---|
| glycolysis | GO:0006096 |  | 1.401E-05 | 0.01074418 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 0.01346938 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.343E-05 | 0.0107755 |
|---|
| glucose catabolic process | GO:0006007 |  | 2.607E-05 | 9.992E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 3.121E-05 | 0.01025573 |
|---|
| regulation of embryonic development | GO:0045995 |  | 3.121E-05 | 8.974E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.764E-05 | 9.619E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.953E-05 | 9.091E-03 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 4.011E-05 | 8.386E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.138E-06 | 0.01255249 |
|---|
| glycolysis | GO:0006096 |  | 5.165E-06 | 6.309E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 7.704E-06 | 6.273E-03 |
|---|
| glucose catabolic process | GO:0006007 |  | 9.386E-06 | 5.733E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.437E-05 | 7.02E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.437E-05 | 5.85E-03 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.687E-05 | 5.888E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.687E-05 | 5.152E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.763E-05 | 4.785E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 1.846E-05 | 4.511E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 2.004E-05 | 4.45E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 4.706E-06 | 0.01132206 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.087E-06 | 6.12E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.087E-06 | 4.08E-03 |
|---|
| glucose catabolic process | GO:0006007 |  | 8.659E-06 | 5.208E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.422E-05 | 6.845E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.422E-05 | 5.704E-03 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.572E-05 | 5.404E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.644E-05 | 4.944E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.718E-05 | 4.593E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 1.828E-05 | 4.398E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.953E-05 | 4.271E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.138E-06 | 0.01255249 |
|---|
| glycolysis | GO:0006096 |  | 5.165E-06 | 6.309E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 7.704E-06 | 6.273E-03 |
|---|
| glucose catabolic process | GO:0006007 |  | 9.386E-06 | 5.733E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.437E-05 | 7.02E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.437E-05 | 5.85E-03 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.687E-05 | 5.888E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.687E-05 | 5.152E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.763E-05 | 4.785E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 1.846E-05 | 4.511E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 2.004E-05 | 4.45E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 4.706E-06 | 0.01132206 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.087E-06 | 6.12E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.087E-06 | 4.08E-03 |
|---|
| glucose catabolic process | GO:0006007 |  | 8.659E-06 | 5.208E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.422E-05 | 6.845E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.422E-05 | 5.704E-03 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.572E-05 | 5.404E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.644E-05 | 4.944E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.718E-05 | 4.593E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 1.828E-05 | 4.398E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.953E-05 | 4.271E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.314E-06 | 0.01183308 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.969E-06 | 8.872E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 6.954E-06 | 8.278E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 6.954E-06 | 6.208E-03 |
|---|
| laminin-1 complex | GO:0005606 |  | 6.954E-06 | 4.967E-03 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 9.269E-06 | 5.517E-03 |
|---|
| T cell homeostasis | GO:0043029 |  | 9.269E-06 | 4.729E-03 |
|---|
| regulation of cell adhesion | GO:0030155 |  | 1.172E-05 | 5.23E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.191E-05 | 4.727E-03 |
|---|
| glucose transmembrane transporter activity | GO:0005355 |  | 1.191E-05 | 4.255E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.362E-05 | 4.423E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 3.136E-06 | 0.0114404 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 3.136E-06 | 5.72E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.703E-06 | 5.718E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 4.703E-06 | 4.289E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 4.703E-06 | 3.431E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 5.601E-06 | 3.406E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 6.463E-06 | 3.368E-03 |
|---|
| laminin-1 complex | GO:0005606 |  | 6.582E-06 | 3.001E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 7.084E-06 | 2.871E-03 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 7.408E-06 | 2.702E-03 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 8.086E-06 | 2.682E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 4.706E-06 | 0.01132206 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.087E-06 | 6.12E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.087E-06 | 4.08E-03 |
|---|
| glucose catabolic process | GO:0006007 |  | 8.659E-06 | 5.208E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.422E-05 | 6.845E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.422E-05 | 5.704E-03 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.572E-05 | 5.404E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.644E-05 | 4.944E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.718E-05 | 4.593E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 1.828E-05 | 4.398E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.953E-05 | 4.271E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.117E-05 | 0.02568926 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.117E-05 | 0.01284463 |
|---|
| glycolysis | GO:0006096 |  | 1.401E-05 | 0.01074418 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 0.01346938 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.343E-05 | 0.0107755 |
|---|
| glucose catabolic process | GO:0006007 |  | 2.607E-05 | 9.992E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 3.121E-05 | 0.01025573 |
|---|
| regulation of embryonic development | GO:0045995 |  | 3.121E-05 | 8.974E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.764E-05 | 9.619E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.953E-05 | 9.091E-03 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 4.011E-05 | 8.386E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.117E-05 | 0.02568926 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.117E-05 | 0.01284463 |
|---|
| glycolysis | GO:0006096 |  | 1.401E-05 | 0.01074418 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 0.01346938 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.343E-05 | 0.0107755 |
|---|
| glucose catabolic process | GO:0006007 |  | 2.607E-05 | 9.992E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 3.121E-05 | 0.01025573 |
|---|
| regulation of embryonic development | GO:0045995 |  | 3.121E-05 | 8.974E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.764E-05 | 9.619E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.953E-05 | 9.091E-03 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 4.011E-05 | 8.386E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.117E-05 | 0.02568926 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.117E-05 | 0.01284463 |
|---|
| glycolysis | GO:0006096 |  | 1.401E-05 | 0.01074418 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 0.01346938 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.343E-05 | 0.0107755 |
|---|
| glucose catabolic process | GO:0006007 |  | 2.607E-05 | 9.992E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 3.121E-05 | 0.01025573 |
|---|
| regulation of embryonic development | GO:0045995 |  | 3.121E-05 | 8.974E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.764E-05 | 9.619E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.953E-05 | 9.091E-03 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 4.011E-05 | 8.386E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.117E-05 | 0.02568926 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.117E-05 | 0.01284463 |
|---|
| glycolysis | GO:0006096 |  | 1.401E-05 | 0.01074418 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 0.01346938 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.343E-05 | 0.0107755 |
|---|
| glucose catabolic process | GO:0006007 |  | 2.607E-05 | 9.992E-03 |
|---|
| positive regulation of Ras GTPase activity | GO:0032320 |  | 3.121E-05 | 0.01025573 |
|---|
| regulation of embryonic development | GO:0045995 |  | 3.121E-05 | 8.974E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.764E-05 | 9.619E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.953E-05 | 9.091E-03 |
|---|
| positive regulation of GTPase activity | GO:0043547 |  | 4.011E-05 | 8.386E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 1.887E-07 | 4.478E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 4.31E-07 | 5.114E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 8.049E-07 | 6.367E-04 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 1.182E-06 | 7.014E-04 |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.844E-06 | 8.751E-04 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 3.372E-06 | 1.334E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.06E-05 | 3.593E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.589E-05 | 4.713E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.223E-05 | 5.861E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.962E-05 | 7.029E-03 |
|---|
| regulation of embryonic development | GO:0045995 |  | 3.806E-05 | 8.21E-03 |
|---|



