Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6311-8-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2515 | 5.553e-03 | 6.606e-03 | 6.327e-02 |
|---|
| IPC-NIBC-129 | 0.2753 | 1.509e-02 | 1.748e-02 | 1.767e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2767 | 5.929e-03 | 9.620e-03 | 1.904e-01 |
|---|
| Loi_GPL570 | 0.2674 | 4.347e-02 | 4.313e-02 | 2.296e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1291 | 4.000e-06 | 3.200e-05 | 2.200e-05 |
|---|
| Parker_GPL1390 | 0.3441 | 4.749e-02 | 4.413e-02 | 3.662e-01 |
|---|
| Parker_GPL887 | 0.2023 | 3.508e-01 | 3.855e-01 | 3.628e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3625 | 9.479e-01 | 9.501e-01 | 1.000e+00 |
|---|
| Schmidt | 0.2505 | 4.821e-03 | 7.102e-03 | 7.296e-02 |
|---|
| Sotiriou | 0.3595 | 4.723e-03 | 1.614e-03 | 9.784e-02 |
|---|
| Wang | 0.2162 | 1.807e-01 | 1.373e-01 | 5.272e-01 |
|---|
| Zhang | 0.0170 | 6.206e-02 | 3.699e-02 | 1.558e-01 |
|---|
| Zhou | 0.3745 | 5.987e-02 | 6.318e-02 | 3.714e-01 |
|---|
Expression data for subnetwork 6311-8-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
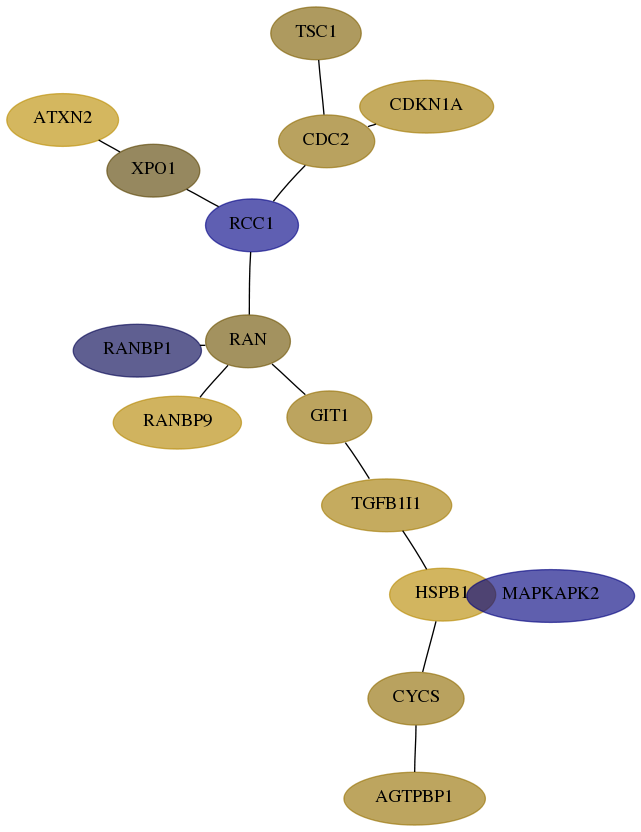
Score for each gene in subnetwork 6311-8-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| agtpbp1 |   | 11 | 21 | 17 | 13 | 0.086 | 0.219 | 0.009 | 0.181 | 0.071 | 0.078 | 0.296 | 0.026 | 0.110 | 0.151 | 0.103 | 0.064 | 0.226 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| cycs |   | 10 | 25 | 17 | 14 | 0.073 | 0.206 | 0.080 | 0.094 | 0.117 | -0.056 | 0.202 | 0.272 | 0.157 | 0.077 | 0.190 | 0.135 | 0.234 |
|---|
| atxn2 |   | 1 | 164 | 149 | 160 | 0.205 | -0.097 | -0.006 | 0.005 | 0.122 | 0.131 | -0.030 | 0.037 | -0.188 | 0.215 | -0.053 | 0.087 | 0.198 |
|---|
| ranbp1 |   | 4 | 60 | 100 | 91 | -0.016 | 0.220 | 0.154 | 0.205 | 0.114 | 0.234 | 0.180 | 0.253 | 0.217 | 0.082 | -0.006 | -0.003 | 0.136 |
|---|
| hspb1 |   | 19 | 11 | 17 | 11 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | -0.026 | -0.065 | 0.289 | -0.021 | 0.201 | 0.125 | -0.077 | 0.290 |
|---|
| ran |   | 4 | 60 | 100 | 91 | 0.032 | 0.181 | 0.147 | 0.083 | 0.166 | 0.250 | -0.287 | 0.247 | 0.127 | 0.178 | 0.052 | -0.025 | 0.254 |
|---|
| tsc1 |   | 47 | 5 | 17 | 8 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.043 | -0.007 | 0.207 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| ranbp9 |   | 4 | 60 | 100 | 91 | 0.172 | 0.207 | 0.012 | 0.253 | 0.126 | 0.186 | -0.113 | 0.148 | 0.143 | 0.127 | 0.205 | 0.021 | 0.164 |
|---|
| mapkapk2 |   | 13 | 19 | 17 | 12 | -0.050 | 0.102 | 0.156 | -0.116 | 0.090 | 0.168 | 0.303 | 0.074 | 0.171 | 0.117 | 0.085 | -0.124 | 0.312 |
|---|
| xpo1 |   | 3 | 79 | 100 | 95 | 0.020 | 0.153 | 0.081 | 0.020 | 0.092 | 0.151 | -0.080 | 0.101 | 0.194 | 0.104 | 0.140 | 0.090 | 0.317 |
|---|
| tgfb1i1 |   | 3 | 79 | 100 | 95 | 0.114 | 0.089 | -0.023 | -0.034 | -0.008 | 0.152 | 0.381 | -0.183 | -0.102 | -0.076 | 0.198 | -0.065 | -0.057 |
|---|
| rcc1 |   | 4 | 60 | 100 | 91 | -0.056 | 0.114 | 0.136 | 0.093 | 0.114 | 0.146 | 0.325 | 0.167 | 0.113 | 0.063 | -0.108 | 0.072 | 0.253 |
|---|
| git1 |   | 5 | 50 | 78 | 70 | 0.082 | -0.027 | 0.097 | 0.051 | 0.077 | 0.074 | -0.099 | 0.138 | 0.087 | 0.097 | -0.065 | -0.065 | 0.114 |
|---|
GO Enrichment output for subnetwork 6311-8-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle organization | GO:0007051 |  | 8.691E-08 | 1.999E-04 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 1.118E-07 | 1.286E-04 |
|---|
| establishment of RNA localization | GO:0051236 |  | 1.414E-07 | 1.084E-04 |
|---|
| RNA localization | GO:0006403 |  | 1.674E-07 | 9.623E-05 |
|---|
| nucleobase. nucleoside. nucleotide and nucleic acid transport | GO:0015931 |  | 2.189E-07 | 1.007E-04 |
|---|
| mitotic spindle organization | GO:0007052 |  | 4.23E-07 | 1.622E-04 |
|---|
| nuclear export | GO:0051168 |  | 6.065E-07 | 1.993E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 1.429E-06 | 4.108E-04 |
|---|
| regulation of translation | GO:0006417 |  | 1.163E-05 | 2.971E-03 |
|---|
| RNA export from nucleus | GO:0006405 |  | 1.37E-05 | 3.151E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.37E-05 | 2.865E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle organization | GO:0007051 |  | 3.498E-08 | 8.546E-05 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 5.793E-08 | 7.077E-05 |
|---|
| mitotic spindle organization | GO:0007052 |  | 2.269E-07 | 1.848E-04 |
|---|
| nuclear export | GO:0051168 |  | 4.34E-07 | 2.65E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 6.99E-07 | 3.415E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 9.946E-07 | 4.05E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.363E-06 | 4.757E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.577E-06 | 4.817E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 2.07E-06 | 5.619E-04 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 7.873E-06 | 1.923E-03 |
|---|
| nucleus organization | GO:0006997 |  | 9.989E-06 | 2.219E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle organization | GO:0007051 |  | 6.035E-08 | 1.452E-04 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 8.747E-08 | 1.052E-04 |
|---|
| mitotic spindle organization | GO:0007052 |  | 3.425E-07 | 2.747E-04 |
|---|
| nuclear export | GO:0051168 |  | 5.613E-07 | 3.376E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.054E-06 | 5.073E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 1.5E-06 | 6.015E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.055E-06 | 7.064E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 2.378E-06 | 7.152E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 3.12E-06 | 8.341E-04 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.09E-05 | 2.622E-03 |
|---|
| RNA export from nucleus | GO:0006405 |  | 1.185E-05 | 2.592E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle organization | GO:0007051 |  | 3.498E-08 | 8.546E-05 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 5.793E-08 | 7.077E-05 |
|---|
| mitotic spindle organization | GO:0007052 |  | 2.269E-07 | 1.848E-04 |
|---|
| nuclear export | GO:0051168 |  | 4.34E-07 | 2.65E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 6.99E-07 | 3.415E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 9.946E-07 | 4.05E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.363E-06 | 4.757E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.577E-06 | 4.817E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 2.07E-06 | 5.619E-04 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 7.873E-06 | 1.923E-03 |
|---|
| nucleus organization | GO:0006997 |  | 9.989E-06 | 2.219E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle organization | GO:0007051 |  | 6.035E-08 | 1.452E-04 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 8.747E-08 | 1.052E-04 |
|---|
| mitotic spindle organization | GO:0007052 |  | 3.425E-07 | 2.747E-04 |
|---|
| nuclear export | GO:0051168 |  | 5.613E-07 | 3.376E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.054E-06 | 5.073E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 1.5E-06 | 6.015E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.055E-06 | 7.064E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 2.378E-06 | 7.152E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 3.12E-06 | 8.341E-04 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.09E-05 | 2.622E-03 |
|---|
| RNA export from nucleus | GO:0006405 |  | 1.185E-05 | 2.592E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| establishment of RNA localization | GO:0051236 |  | 5.766E-09 | 2.059E-05 |
|---|
| RNA localization | GO:0006403 |  | 6.594E-09 | 1.177E-05 |
|---|
| spindle organization | GO:0007051 |  | 1.195E-08 | 1.422E-05 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 1.227E-08 | 1.095E-05 |
|---|
| nucleobase. nucleoside. nucleotide and nucleic acid transport | GO:0015931 |  | 1.377E-08 | 9.833E-06 |
|---|
| nuclear export | GO:0051168 |  | 4.57E-08 | 2.72E-05 |
|---|
| Ran GTPase binding | GO:0008536 |  | 8.163E-08 | 4.164E-05 |
|---|
| mitotic spindle organization | GO:0007052 |  | 1.495E-07 | 6.673E-05 |
|---|
| protein export from nucleus | GO:0006611 |  | 3.087E-07 | 1.225E-04 |
|---|
| androgen receptor binding | GO:0050681 |  | 1.196E-06 | 4.271E-04 |
|---|
| RNA export from nucleus | GO:0006405 |  | 1.552E-06 | 5.037E-04 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| establishment of RNA localization | GO:0051236 |  | 8.48E-09 | 3.093E-05 |
|---|
| RNA localization | GO:0006403 |  | 9.661E-09 | 1.762E-05 |
|---|
| spindle organization | GO:0007051 |  | 1.035E-08 | 1.259E-05 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 1.577E-08 | 1.438E-05 |
|---|
| nucleobase. nucleoside. nucleotide and nucleic acid transport | GO:0015931 |  | 1.98E-08 | 1.444E-05 |
|---|
| Ran GTPase binding | GO:0008536 |  | 6.247E-08 | 3.798E-05 |
|---|
| nuclear export | GO:0051168 |  | 6.545E-08 | 3.411E-05 |
|---|
| mitotic spindle organization | GO:0007052 |  | 1.633E-07 | 7.448E-05 |
|---|
| protein export from nucleus | GO:0006611 |  | 2.699E-07 | 1.094E-04 |
|---|
| androgen receptor binding | GO:0050681 |  | 1.492E-06 | 5.442E-04 |
|---|
| RNA export from nucleus | GO:0006405 |  | 2.152E-06 | 7.137E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle organization | GO:0007051 |  | 6.035E-08 | 1.452E-04 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 8.747E-08 | 1.052E-04 |
|---|
| mitotic spindle organization | GO:0007052 |  | 3.425E-07 | 2.747E-04 |
|---|
| nuclear export | GO:0051168 |  | 5.613E-07 | 3.376E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.054E-06 | 5.073E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 1.5E-06 | 6.015E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.055E-06 | 7.064E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 2.378E-06 | 7.152E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 3.12E-06 | 8.341E-04 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.09E-05 | 2.622E-03 |
|---|
| RNA export from nucleus | GO:0006405 |  | 1.185E-05 | 2.592E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle organization | GO:0007051 |  | 8.691E-08 | 1.999E-04 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 1.118E-07 | 1.286E-04 |
|---|
| establishment of RNA localization | GO:0051236 |  | 1.414E-07 | 1.084E-04 |
|---|
| RNA localization | GO:0006403 |  | 1.674E-07 | 9.623E-05 |
|---|
| nucleobase. nucleoside. nucleotide and nucleic acid transport | GO:0015931 |  | 2.189E-07 | 1.007E-04 |
|---|
| mitotic spindle organization | GO:0007052 |  | 4.23E-07 | 1.622E-04 |
|---|
| nuclear export | GO:0051168 |  | 6.065E-07 | 1.993E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 1.429E-06 | 4.108E-04 |
|---|
| regulation of translation | GO:0006417 |  | 1.163E-05 | 2.971E-03 |
|---|
| RNA export from nucleus | GO:0006405 |  | 1.37E-05 | 3.151E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.37E-05 | 2.865E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle organization | GO:0007051 |  | 8.691E-08 | 1.999E-04 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 1.118E-07 | 1.286E-04 |
|---|
| establishment of RNA localization | GO:0051236 |  | 1.414E-07 | 1.084E-04 |
|---|
| RNA localization | GO:0006403 |  | 1.674E-07 | 9.623E-05 |
|---|
| nucleobase. nucleoside. nucleotide and nucleic acid transport | GO:0015931 |  | 2.189E-07 | 1.007E-04 |
|---|
| mitotic spindle organization | GO:0007052 |  | 4.23E-07 | 1.622E-04 |
|---|
| nuclear export | GO:0051168 |  | 6.065E-07 | 1.993E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 1.429E-06 | 4.108E-04 |
|---|
| regulation of translation | GO:0006417 |  | 1.163E-05 | 2.971E-03 |
|---|
| RNA export from nucleus | GO:0006405 |  | 1.37E-05 | 3.151E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.37E-05 | 2.865E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle organization | GO:0007051 |  | 8.691E-08 | 1.999E-04 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 1.118E-07 | 1.286E-04 |
|---|
| establishment of RNA localization | GO:0051236 |  | 1.414E-07 | 1.084E-04 |
|---|
| RNA localization | GO:0006403 |  | 1.674E-07 | 9.623E-05 |
|---|
| nucleobase. nucleoside. nucleotide and nucleic acid transport | GO:0015931 |  | 2.189E-07 | 1.007E-04 |
|---|
| mitotic spindle organization | GO:0007052 |  | 4.23E-07 | 1.622E-04 |
|---|
| nuclear export | GO:0051168 |  | 6.065E-07 | 1.993E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 1.429E-06 | 4.108E-04 |
|---|
| regulation of translation | GO:0006417 |  | 1.163E-05 | 2.971E-03 |
|---|
| RNA export from nucleus | GO:0006405 |  | 1.37E-05 | 3.151E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.37E-05 | 2.865E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle organization | GO:0007051 |  | 8.691E-08 | 1.999E-04 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 1.118E-07 | 1.286E-04 |
|---|
| establishment of RNA localization | GO:0051236 |  | 1.414E-07 | 1.084E-04 |
|---|
| RNA localization | GO:0006403 |  | 1.674E-07 | 9.623E-05 |
|---|
| nucleobase. nucleoside. nucleotide and nucleic acid transport | GO:0015931 |  | 2.189E-07 | 1.007E-04 |
|---|
| mitotic spindle organization | GO:0007052 |  | 4.23E-07 | 1.622E-04 |
|---|
| nuclear export | GO:0051168 |  | 6.065E-07 | 1.993E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 1.429E-06 | 4.108E-04 |
|---|
| regulation of translation | GO:0006417 |  | 1.163E-05 | 2.971E-03 |
|---|
| RNA export from nucleus | GO:0006405 |  | 1.37E-05 | 3.151E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.37E-05 | 2.865E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 5.352E-08 | 1.27E-04 |
|---|
| spindle organization | GO:0007051 |  | 5.438E-08 | 6.453E-05 |
|---|
| mitotic spindle organization | GO:0007052 |  | 2.952E-07 | 2.335E-04 |
|---|
| nuclear export | GO:0051168 |  | 5.154E-07 | 3.058E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 9.09E-07 | 4.314E-04 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.023E-05 | 4.047E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.277E-05 | 4.328E-03 |
|---|
| RNA export from nucleus | GO:0006405 |  | 1.399E-05 | 4.151E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.735E-05 | 4.574E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.914E-05 | 4.541E-03 |
|---|
| microtubule nucleation | GO:0007020 |  | 1.914E-05 | 4.128E-03 |
|---|



