Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 60598-7-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2178 | 2.666e-02 | 3.198e-02 | 2.011e-01 |
|---|
| IPC-NIBC-129 | 0.3107 | 7.951e-02 | 1.038e-01 | 5.987e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2004 | 1.292e-03 | 2.445e-03 | 6.591e-02 |
|---|
| Loi_GPL570 | 0.3379 | 2.657e-02 | 2.633e-02 | 1.568e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1384 | 0.000e+00 | 0.000e+00 | 0.000e+00 |
|---|
| Parker_GPL1390 | 0.2243 | 7.657e-02 | 7.452e-02 | 4.735e-01 |
|---|
| Parker_GPL887 | 0.0273 | 9.263e-01 | 9.345e-01 | 9.601e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3814 | 6.854e-02 | 4.717e-02 | 4.065e-01 |
|---|
| Schmidt | 0.3192 | 1.368e-02 | 1.928e-02 | 1.659e-01 |
|---|
| Sotiriou | 0.2960 | 1.727e-03 | 4.530e-04 | 4.758e-02 |
|---|
| Wang | 0.0551 | 1.562e-01 | 1.158e-01 | 4.836e-01 |
|---|
| Zhang | 0.3076 | 5.738e-01 | 5.263e-01 | 8.603e-01 |
|---|
| Zhou | 0.4351 | 4.995e-02 | 5.178e-02 | 3.168e-01 |
|---|
Expression data for subnetwork 60598-7-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
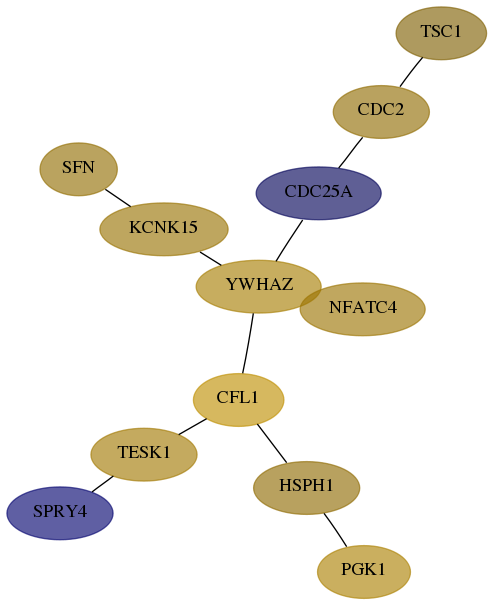
Score for each gene in subnetwork 60598-7-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| kcnk15 |   | 1 | 164 | 191 | 199 | 0.088 | -0.091 | -0.068 | 0.220 | 0.076 | -0.136 | 0.026 | 0.083 | 0.037 | 0.154 | 0.087 | 0.098 | 0.140 |
|---|
| nfatc4 |   | 8 | 33 | 31 | 25 | 0.094 | -0.056 | -0.021 | 0.204 | 0.079 | -0.008 | 0.243 | 0.060 | 0.077 | -0.012 | 0.080 | 0.131 | 0.154 |
|---|
| ywhaz |   | 11 | 21 | 31 | 20 | 0.124 | 0.089 | 0.087 | 0.200 | 0.141 | 0.110 | 0.228 | 0.199 | 0.056 | 0.123 | -0.026 | 0.103 | 0.213 |
|---|
| pgk1 |   | 5 | 50 | 53 | 47 | 0.141 | 0.293 | 0.135 | 0.197 | 0.170 | 0.170 | 0.120 | 0.310 | 0.201 | 0.127 | 0.110 | 0.054 | 0.141 |
|---|
| cfl1 |   | 2 | 104 | 53 | 62 | 0.216 | 0.134 | 0.083 | -0.084 | 0.142 | 0.251 | -0.466 | 0.229 | 0.174 | 0.188 | 0.021 | -0.025 | 0.215 |
|---|
| tsc1 |   | 47 | 5 | 17 | 8 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.043 | -0.007 | 0.207 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| hsph1 |   | 4 | 60 | 53 | 50 | 0.071 | 0.260 | 0.138 | 0.050 | 0.148 | 0.197 | -0.116 | 0.161 | 0.143 | 0.132 | 0.169 | 0.094 | 0.357 |
|---|
| cdc25a |   | 9 | 27 | 109 | 90 | -0.019 | 0.221 | 0.093 | 0.158 | 0.120 | -0.061 | 0.406 | 0.207 | 0.211 | 0.016 | 0.069 | 0.183 | 0.264 |
|---|
| sfn |   | 50 | 4 | 29 | 17 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.025 | 0.079 | 0.181 |
|---|
| tesk1 |   | 2 | 104 | 53 | 62 | 0.110 | 0.169 | 0.161 | 0.179 | 0.096 | 0.138 | -0.154 | 0.245 | 0.155 | 0.092 | 0.022 | 0.029 | 0.200 |
|---|
| spry4 |   | 2 | 104 | 53 | 62 | -0.033 | 0.158 | -0.001 | -0.039 | 0.103 | 0.074 | -0.328 | 0.022 | 0.197 | 0.156 | -0.057 | 0.184 | 0.211 |
|---|
GO Enrichment output for subnetwork 60598-7-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 7.199E-10 | 1.656E-06 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.806E-09 | 2.077E-06 |
|---|
| polyol catabolic process | GO:0046174 |  | 1.986E-08 | 1.522E-05 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.986E-08 | 1.142E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.662E-07 | 7.647E-05 |
|---|
| glycolysis | GO:0006096 |  | 2.984E-07 | 1.144E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.26E-07 | 1.071E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 6.892E-07 | 1.981E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.102E-06 | 2.815E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.373E-06 | 3.157E-04 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 2.086E-06 | 4.362E-04 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.265E-10 | 3.09E-07 |
|---|
| polyol catabolic process | GO:0046174 |  | 5.999E-09 | 7.328E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 5.999E-09 | 4.885E-06 |
|---|
| neural plate morphogenesis | GO:0001839 |  | 5.999E-09 | 3.664E-06 |
|---|
| neural plate development | GO:0001840 |  | 1.199E-08 | 5.859E-06 |
|---|
| glycolysis | GO:0006096 |  | 7.527E-08 | 3.065E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 9.864E-08 | 3.443E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 1.314E-07 | 4.014E-05 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.684E-07 | 4.571E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.708E-07 | 4.172E-05 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.338E-07 | 7.413E-05 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 9.983E-11 | 2.402E-07 |
|---|
| polyol catabolic process | GO:0046174 |  | 5.705E-09 | 6.863E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 5.705E-09 | 4.575E-06 |
|---|
| glycolysis | GO:0006096 |  | 6.345E-08 | 3.817E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 9.381E-08 | 4.514E-05 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.444E-07 | 5.79E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.624E-07 | 5.582E-05 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.175E-07 | 9.548E-05 |
|---|
| hexose catabolic process | GO:0019320 |  | 3.223E-07 | 8.615E-05 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 5.366E-07 | 1.291E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.142E-06 | 2.498E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.265E-10 | 3.09E-07 |
|---|
| polyol catabolic process | GO:0046174 |  | 5.999E-09 | 7.328E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 5.999E-09 | 4.885E-06 |
|---|
| neural plate morphogenesis | GO:0001839 |  | 5.999E-09 | 3.664E-06 |
|---|
| neural plate development | GO:0001840 |  | 1.199E-08 | 5.859E-06 |
|---|
| glycolysis | GO:0006096 |  | 7.527E-08 | 3.065E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 9.864E-08 | 3.443E-05 |
|---|
| establishment of cell polarity | GO:0030010 |  | 1.314E-07 | 4.014E-05 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.684E-07 | 4.571E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.708E-07 | 4.172E-05 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.338E-07 | 7.413E-05 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 9.983E-11 | 2.402E-07 |
|---|
| polyol catabolic process | GO:0046174 |  | 5.705E-09 | 6.863E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 5.705E-09 | 4.575E-06 |
|---|
| glycolysis | GO:0006096 |  | 6.345E-08 | 3.817E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 9.381E-08 | 4.514E-05 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.444E-07 | 5.79E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.624E-07 | 5.582E-05 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.175E-07 | 9.548E-05 |
|---|
| hexose catabolic process | GO:0019320 |  | 3.223E-07 | 8.615E-05 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 5.366E-07 | 1.291E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.142E-06 | 2.498E-04 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| neural plate development | GO:0001840 |  | 4.142E-06 | 0.01478934 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 4.142E-06 | 7.395E-03 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 4.142E-06 | 4.93E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.339E-06 | 3.874E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 7.937E-06 | 5.669E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 8.338E-06 | 4.962E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 8.691E-06 | 4.434E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 8.691E-06 | 3.879E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 9.619E-06 | 3.817E-03 |
|---|
| regulation of actin cytoskeleton organization | GO:0032956 |  | 1.102E-05 | 3.937E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 1.158E-05 | 3.76E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| neural plate morphogenesis | GO:0001839 |  | 3.763E-06 | 0.01372702 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 3.763E-06 | 6.864E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 3.763E-06 | 4.576E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.119E-06 | 3.756E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 5.642E-06 | 4.117E-03 |
|---|
| neural plate development | GO:0001840 |  | 5.642E-06 | 3.431E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.642E-06 | 2.94E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 8.202E-06 | 3.74E-03 |
|---|
| regulation of actin cytoskeleton organization | GO:0032956 |  | 8.202E-06 | 3.325E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 8.596E-06 | 3.136E-03 |
|---|
| regulation of actin filament-based process | GO:0032970 |  | 9.421E-06 | 3.124E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 9.983E-11 | 2.402E-07 |
|---|
| polyol catabolic process | GO:0046174 |  | 5.705E-09 | 6.863E-06 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 5.705E-09 | 4.575E-06 |
|---|
| glycolysis | GO:0006096 |  | 6.345E-08 | 3.817E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 9.381E-08 | 4.514E-05 |
|---|
| glucose catabolic process | GO:0006007 |  | 1.444E-07 | 5.79E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.624E-07 | 5.582E-05 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.175E-07 | 9.548E-05 |
|---|
| hexose catabolic process | GO:0019320 |  | 3.223E-07 | 8.615E-05 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 5.366E-07 | 1.291E-04 |
|---|
| alditol metabolic process | GO:0019400 |  | 1.142E-06 | 2.498E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 7.199E-10 | 1.656E-06 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.806E-09 | 2.077E-06 |
|---|
| polyol catabolic process | GO:0046174 |  | 1.986E-08 | 1.522E-05 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.986E-08 | 1.142E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.662E-07 | 7.647E-05 |
|---|
| glycolysis | GO:0006096 |  | 2.984E-07 | 1.144E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.26E-07 | 1.071E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 6.892E-07 | 1.981E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.102E-06 | 2.815E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.373E-06 | 3.157E-04 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 2.086E-06 | 4.362E-04 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 7.199E-10 | 1.656E-06 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.806E-09 | 2.077E-06 |
|---|
| polyol catabolic process | GO:0046174 |  | 1.986E-08 | 1.522E-05 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.986E-08 | 1.142E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.662E-07 | 7.647E-05 |
|---|
| glycolysis | GO:0006096 |  | 2.984E-07 | 1.144E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.26E-07 | 1.071E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 6.892E-07 | 1.981E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.102E-06 | 2.815E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.373E-06 | 3.157E-04 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 2.086E-06 | 4.362E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 7.199E-10 | 1.656E-06 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.806E-09 | 2.077E-06 |
|---|
| polyol catabolic process | GO:0046174 |  | 1.986E-08 | 1.522E-05 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.986E-08 | 1.142E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.662E-07 | 7.647E-05 |
|---|
| glycolysis | GO:0006096 |  | 2.984E-07 | 1.144E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.26E-07 | 1.071E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 6.892E-07 | 1.981E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.102E-06 | 2.815E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.373E-06 | 3.157E-04 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 2.086E-06 | 4.362E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 7.199E-10 | 1.656E-06 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.806E-09 | 2.077E-06 |
|---|
| polyol catabolic process | GO:0046174 |  | 1.986E-08 | 1.522E-05 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.986E-08 | 1.142E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.662E-07 | 7.647E-05 |
|---|
| glycolysis | GO:0006096 |  | 2.984E-07 | 1.144E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.26E-07 | 1.071E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 6.892E-07 | 1.981E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.102E-06 | 2.815E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.373E-06 | 3.157E-04 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 2.086E-06 | 4.362E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| alcohol catabolic process | GO:0046164 |  | 9.08E-10 | 2.155E-06 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 2.276E-09 | 2.7E-06 |
|---|
| polyol catabolic process | GO:0046174 |  | 1.752E-08 | 1.386E-05 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.752E-08 | 1.039E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 2.093E-07 | 9.935E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 3.831E-07 | 1.515E-04 |
|---|
| glycolysis | GO:0006096 |  | 3.959E-07 | 1.342E-04 |
|---|
| glucose catabolic process | GO:0006007 |  | 9.028E-07 | 2.678E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.179E-06 | 3.109E-04 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.683E-06 | 3.994E-04 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 2.47E-06 | 5.328E-04 |
|---|



