Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 60312-7-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1535 | 6.175e-03 | 7.353e-03 | 6.868e-02 |
|---|
| IPC-NIBC-129 | 0.2504 | 1.367e-02 | 1.570e-02 | 1.617e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2716 | 1.214e-02 | 1.833e-02 | 2.952e-01 |
|---|
| Loi_GPL570 | 0.2339 | 6.112e-02 | 6.069e-02 | 2.949e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1717 | 1.000e-06 | 6.000e-06 | 1.000e-06 |
|---|
| Parker_GPL1390 | 0.3511 | 1.858e-01 | 1.943e-01 | 7.096e-01 |
|---|
| Parker_GPL887 | 0.1074 | 6.230e-01 | 6.524e-01 | 6.731e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3797 | 1.418e-01 | 1.097e-01 | 6.316e-01 |
|---|
| Schmidt | 0.2768 | 4.446e-03 | 6.571e-03 | 6.823e-02 |
|---|
| Sotiriou | 0.3644 | 1.800e-03 | 4.780e-04 | 4.905e-02 |
|---|
| Wang | 0.1612 | 9.170e-02 | 6.215e-02 | 3.453e-01 |
|---|
| Zhang | 0.2369 | 1.369e-01 | 9.539e-02 | 3.156e-01 |
|---|
| Zhou | 0.4326 | 5.078e-02 | 5.273e-02 | 3.216e-01 |
|---|
Expression data for subnetwork 60312-7-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
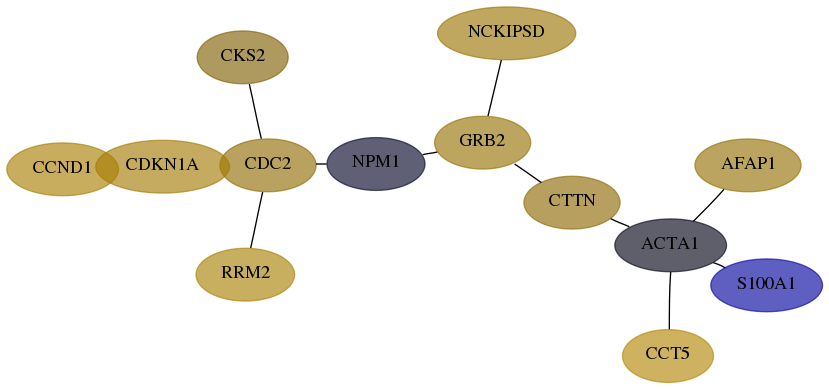
Score for each gene in subnetwork 60312-7-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| afap1 |   | 1 | 164 | 87 | 105 | 0.080 | 0.112 | 0.020 | 0.064 | 0.063 | 0.171 | -0.283 | 0.200 | -0.021 | 0.119 | 0.078 | 0.063 | 0.241 |
|---|
| rrm2 |   | 21 | 10 | 44 | 26 | 0.131 | 0.207 | 0.227 | 0.187 | 0.161 | 0.178 | 0.137 | 0.303 | 0.190 | 0.219 | 0.117 | 0.129 | 0.302 |
|---|
| cttn |   | 9 | 27 | 44 | 37 | 0.067 | 0.065 | 0.123 | 0.238 | 0.126 | 0.263 | -0.172 | 0.216 | 0.069 | 0.115 | 0.129 | 0.025 | 0.279 |
|---|
| s100a1 |   | 2 | 104 | 44 | 52 | -0.100 | 0.147 | 0.022 | -0.057 | 0.048 | 0.011 | 0.134 | 0.085 | 0.103 | 0.032 | 0.027 | 0.014 | -0.049 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| cct5 |   | 2 | 104 | 44 | 52 | 0.165 | 0.258 | 0.182 | 0.155 | 0.209 | 0.212 | 0.257 | 0.353 | 0.143 | 0.204 | 0.106 | 0.039 | 0.289 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| cks2 |   | 44 | 6 | 73 | 54 | 0.050 | 0.233 | 0.170 | 0.103 | 0.186 | 0.172 | 0.093 | 0.167 | 0.263 | 0.151 | 0.145 | 0.284 | 0.383 |
|---|
| nckipsd |   | 3 | 79 | 44 | 44 | 0.094 | 0.086 | 0.221 | -0.049 | 0.109 | -0.007 | 0.119 | 0.121 | 0.006 | 0.126 | 0.073 | 0.054 | 0.339 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| acta1 |   | 6 | 43 | 44 | 40 | -0.003 | 0.098 | -0.008 | -0.030 | 0.119 | 0.174 | 0.376 | 0.024 | 0.182 | 0.214 | -0.013 | 0.048 | 0.201 |
|---|
| npm1 |   | 4 | 60 | 44 | 41 | -0.005 | 0.172 | 0.085 | 0.197 | 0.093 | 0.221 | -0.227 | -0.050 | 0.065 | 0.169 | -0.000 | -0.086 | 0.199 |
|---|
| grb2 |   | 9 | 27 | 1 | 4 | 0.084 | -0.013 | 0.086 | -0.031 | 0.162 | -0.038 | 0.003 | 0.065 | 0.158 | 0.127 | -0.097 | 0.194 | -0.005 |
|---|
GO Enrichment output for subnetwork 60312-7-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.137E-11 | 2.615E-08 |
|---|
| ribosome assembly | GO:0042255 |  | 7.465E-10 | 8.585E-07 |
|---|
| centrosome cycle | GO:0007098 |  | 3.077E-09 | 2.359E-06 |
|---|
| cell aging | GO:0007569 |  | 1.342E-08 | 7.718E-06 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 1.639E-08 | 7.54E-06 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 3.338E-08 | 1.28E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.968E-08 | 1.304E-05 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 6.098E-08 | 1.753E-05 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 6.098E-08 | 1.558E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.938E-08 | 1.596E-05 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 7.137E-08 | 1.492E-05 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.319E-12 | 3.222E-09 |
|---|
| ribosome assembly | GO:0042255 |  | 2.628E-10 | 3.211E-07 |
|---|
| centrosome cycle | GO:0007098 |  | 8.018E-10 | 6.529E-07 |
|---|
| cell aging | GO:0007569 |  | 1.913E-09 | 1.168E-06 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 3.901E-09 | 1.906E-06 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 8.189E-09 | 3.334E-06 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 9.354E-09 | 3.265E-06 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 1.714E-08 | 5.233E-06 |
|---|
| positive regulation of transcription factor activity | GO:0051091 |  | 3.192E-08 | 8.665E-06 |
|---|
| positive regulation of DNA binding | GO:0043388 |  | 5.02E-08 | 1.226E-05 |
|---|
| positive regulation of binding | GO:0051099 |  | 6.97E-08 | 1.548E-05 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 2.18E-12 | 5.244E-09 |
|---|
| ribosome assembly | GO:0042255 |  | 3.103E-10 | 3.732E-07 |
|---|
| centrosome cycle | GO:0007098 |  | 1.324E-09 | 1.062E-06 |
|---|
| cell aging | GO:0007569 |  | 2.585E-09 | 1.555E-06 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 6.438E-09 | 3.098E-06 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 1.021E-08 | 4.094E-06 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 1.543E-08 | 5.304E-06 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.994E-08 | 5.998E-06 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 2.243E-08 | 5.996E-06 |
|---|
| positive regulation of transcription factor activity | GO:0051091 |  | 4.775E-08 | 1.149E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.781E-08 | 1.046E-05 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.319E-12 | 3.222E-09 |
|---|
| ribosome assembly | GO:0042255 |  | 2.628E-10 | 3.211E-07 |
|---|
| centrosome cycle | GO:0007098 |  | 8.018E-10 | 6.529E-07 |
|---|
| cell aging | GO:0007569 |  | 1.913E-09 | 1.168E-06 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 3.901E-09 | 1.906E-06 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 8.189E-09 | 3.334E-06 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 9.354E-09 | 3.265E-06 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 1.714E-08 | 5.233E-06 |
|---|
| positive regulation of transcription factor activity | GO:0051091 |  | 3.192E-08 | 8.665E-06 |
|---|
| positive regulation of DNA binding | GO:0043388 |  | 5.02E-08 | 1.226E-05 |
|---|
| positive regulation of binding | GO:0051099 |  | 6.97E-08 | 1.548E-05 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 2.18E-12 | 5.244E-09 |
|---|
| ribosome assembly | GO:0042255 |  | 3.103E-10 | 3.732E-07 |
|---|
| centrosome cycle | GO:0007098 |  | 1.324E-09 | 1.062E-06 |
|---|
| cell aging | GO:0007569 |  | 2.585E-09 | 1.555E-06 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 6.438E-09 | 3.098E-06 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 1.021E-08 | 4.094E-06 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 1.543E-08 | 5.304E-06 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.994E-08 | 5.998E-06 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 2.243E-08 | 5.996E-06 |
|---|
| positive regulation of transcription factor activity | GO:0051091 |  | 4.775E-08 | 1.149E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.781E-08 | 1.046E-05 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 8.297E-11 | 2.963E-07 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 2.306E-08 | 4.118E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.693E-08 | 6.776E-05 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 1.561E-07 | 1.394E-04 |
|---|
| kinase regulator activity | GO:0019207 |  | 3.306E-07 | 2.361E-04 |
|---|
| skeletal myofibril assembly | GO:0014866 |  | 6.073E-06 | 3.614E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.143E-06 | 3.134E-03 |
|---|
| response to UV | GO:0009411 |  | 7.446E-06 | 3.324E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.105E-06 | 3.613E-03 |
|---|
| insulin receptor substrate binding | GO:0043560 |  | 9.105E-06 | 3.251E-03 |
|---|
| oxidoreductase activity. acting on CH or CH2 groups | GO:0016725 |  | 1.274E-05 | 4.136E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 2.659E-10 | 9.699E-07 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 2.325E-08 | 4.241E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.523E-08 | 7.931E-05 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 1.868E-07 | 1.704E-04 |
|---|
| kinase regulator activity | GO:0019207 |  | 3.309E-07 | 2.415E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.039E-06 | 3.063E-03 |
|---|
| response to UV | GO:0009411 |  | 5.788E-06 | 3.016E-03 |
|---|
| muscle thin filament assembly | GO:0030240 |  | 5.984E-06 | 2.729E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 5.984E-06 | 2.426E-03 |
|---|
| epidermal growth factor receptor binding | GO:0005154 |  | 5.984E-06 | 2.183E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 8.973E-06 | 2.976E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 2.18E-12 | 5.244E-09 |
|---|
| ribosome assembly | GO:0042255 |  | 3.103E-10 | 3.732E-07 |
|---|
| centrosome cycle | GO:0007098 |  | 1.324E-09 | 1.062E-06 |
|---|
| cell aging | GO:0007569 |  | 2.585E-09 | 1.555E-06 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 6.438E-09 | 3.098E-06 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 1.021E-08 | 4.094E-06 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 1.543E-08 | 5.304E-06 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.994E-08 | 5.998E-06 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 2.243E-08 | 5.996E-06 |
|---|
| positive regulation of transcription factor activity | GO:0051091 |  | 4.775E-08 | 1.149E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.781E-08 | 1.046E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.137E-11 | 2.615E-08 |
|---|
| ribosome assembly | GO:0042255 |  | 7.465E-10 | 8.585E-07 |
|---|
| centrosome cycle | GO:0007098 |  | 3.077E-09 | 2.359E-06 |
|---|
| cell aging | GO:0007569 |  | 1.342E-08 | 7.718E-06 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 1.639E-08 | 7.54E-06 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 3.338E-08 | 1.28E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.968E-08 | 1.304E-05 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 6.098E-08 | 1.753E-05 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 6.098E-08 | 1.558E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.938E-08 | 1.596E-05 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 7.137E-08 | 1.492E-05 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.137E-11 | 2.615E-08 |
|---|
| ribosome assembly | GO:0042255 |  | 7.465E-10 | 8.585E-07 |
|---|
| centrosome cycle | GO:0007098 |  | 3.077E-09 | 2.359E-06 |
|---|
| cell aging | GO:0007569 |  | 1.342E-08 | 7.718E-06 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 1.639E-08 | 7.54E-06 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 3.338E-08 | 1.28E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.968E-08 | 1.304E-05 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 6.098E-08 | 1.753E-05 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 6.098E-08 | 1.558E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.938E-08 | 1.596E-05 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 7.137E-08 | 1.492E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.137E-11 | 2.615E-08 |
|---|
| ribosome assembly | GO:0042255 |  | 7.465E-10 | 8.585E-07 |
|---|
| centrosome cycle | GO:0007098 |  | 3.077E-09 | 2.359E-06 |
|---|
| cell aging | GO:0007569 |  | 1.342E-08 | 7.718E-06 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 1.639E-08 | 7.54E-06 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 3.338E-08 | 1.28E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.968E-08 | 1.304E-05 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 6.098E-08 | 1.753E-05 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 6.098E-08 | 1.558E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.938E-08 | 1.596E-05 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 7.137E-08 | 1.492E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.137E-11 | 2.615E-08 |
|---|
| ribosome assembly | GO:0042255 |  | 7.465E-10 | 8.585E-07 |
|---|
| centrosome cycle | GO:0007098 |  | 3.077E-09 | 2.359E-06 |
|---|
| cell aging | GO:0007569 |  | 1.342E-08 | 7.718E-06 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 1.639E-08 | 7.54E-06 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 3.338E-08 | 1.28E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.968E-08 | 1.304E-05 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 6.098E-08 | 1.753E-05 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 6.098E-08 | 1.558E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.938E-08 | 1.596E-05 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 7.137E-08 | 1.492E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome duplication | GO:0010824 |  | 6.789E-12 | 1.611E-08 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 2.035E-11 | 2.415E-08 |
|---|
| ribosome assembly | GO:0042255 |  | 1.839E-09 | 1.455E-06 |
|---|
| centrosome cycle | GO:0007098 |  | 2.45E-09 | 1.454E-06 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 1.186E-08 | 5.628E-06 |
|---|
| cell aging | GO:0007569 |  | 1.422E-08 | 5.624E-06 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 3.166E-08 | 1.073E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.714E-08 | 1.398E-05 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 4.783E-08 | 1.261E-05 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 5.352E-08 | 1.27E-05 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 5.438E-08 | 1.173E-05 |
|---|



