Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 5999-7-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2492 | 8.391e-03 | 1.002e-02 | 8.682e-02 |
|---|
| IPC-NIBC-129 | 0.2461 | 1.490e-01 | 1.982e-01 | 7.968e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2568 | 4.752e-03 | 7.884e-03 | 1.648e-01 |
|---|
| Loi_GPL570 | 0.2777 | 6.480e-02 | 6.436e-02 | 3.075e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1503 | 1.110e-04 | 5.430e-04 | 2.470e-03 |
|---|
| Parker_GPL1390 | 0.1773 | 4.912e-02 | 4.580e-02 | 3.732e-01 |
|---|
| Parker_GPL887 | 0.0889 | 6.899e-01 | 7.160e-01 | 7.447e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3613 | 2.681e-01 | 2.289e-01 | 8.343e-01 |
|---|
| Schmidt | 0.2042 | 1.710e-02 | 2.385e-02 | 1.956e-01 |
|---|
| Sotiriou | 0.2823 | 5.063e-03 | 1.762e-03 | 1.027e-01 |
|---|
| Wang | 0.1910 | 1.293e-01 | 9.282e-02 | 4.306e-01 |
|---|
| Zhang | 0.1724 | 8.939e-02 | 5.727e-02 | 2.175e-01 |
|---|
| Zhou | 0.3703 | 6.057e-02 | 6.398e-02 | 3.751e-01 |
|---|
Expression data for subnetwork 5999-7-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
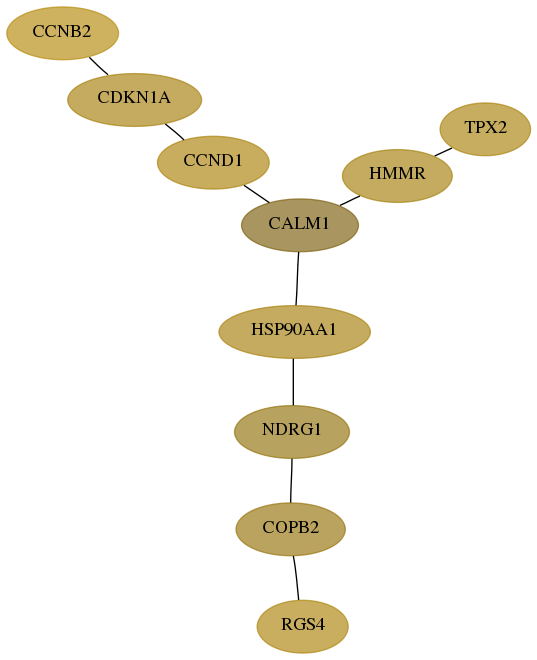
Score for each gene in subnetwork 5999-7-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| ccnb2 |   | 16 | 13 | 153 | 125 | 0.164 | 0.176 | 0.204 | 0.123 | 0.172 | 0.178 | 0.090 | 0.366 | 0.240 | 0.169 | 0.144 | 0.211 | 0.463 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| tpx2 |   | 2 | 104 | 166 | 158 | 0.125 | 0.234 | 0.143 | 0.235 | 0.185 | 0.062 | 0.375 | 0.317 | 0.210 | 0.209 | 0.028 | 0.103 | 0.411 |
|---|
| hsp90aa1 |   | 11 | 21 | 234 | 204 | 0.116 | 0.030 | 0.164 | 0.076 | 0.119 | 0.155 | -0.199 | 0.169 | 0.069 | 0.102 | 0.052 | -0.011 | 0.399 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| ndrg1 |   | 9 | 27 | 250 | 223 | 0.072 | 0.301 | 0.084 | 0.324 | 0.108 | 0.102 | 0.240 | 0.083 | 0.068 | 0.064 | 0.128 | 0.092 | -0.255 |
|---|
| calm1 |   | 4 | 60 | 130 | 115 | 0.038 | 0.023 | 0.171 | 0.277 | 0.113 | 0.034 | -0.199 | 0.007 | 0.171 | 0.112 | 0.087 | 0.162 | 0.171 |
|---|
| hmmr |   | 4 | 60 | 130 | 115 | 0.112 | 0.310 | 0.147 | 0.127 | 0.145 | 0.134 | 0.131 | 0.221 | 0.179 | 0.233 | 0.227 | 0.144 | 0.325 |
|---|
| copb2 |   | 1 | 164 | 387 | 387 | 0.076 | 0.119 | 0.040 | -0.111 | 0.053 | 0.065 | -0.264 | 0.163 | 0.122 | -0.019 | 0.152 | 0.068 | 0.165 |
|---|
| rgs4 |   | 1 | 164 | 387 | 387 | 0.135 | 0.121 | 0.127 | 0.053 | 0.072 | -0.138 | 0.067 | 0.007 | 0.015 | 0.035 | 0.201 | 0.113 | 0.084 |
|---|
GO Enrichment output for subnetwork 5999-7-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.787E-05 | 0.04111132 |
|---|
| protein refolding | GO:0042026 |  | 2.056E-05 | 0.0236386 |
|---|
| response to UV | GO:0009411 |  | 2.194E-05 | 0.01681726 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 0.01653503 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 2.876E-05 | 0.01322803 |
|---|
| mast cell activation | GO:0045576 |  | 2.876E-05 | 0.01102336 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.998E-05 | 9.85E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.831E-05 | 0.01101537 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 3.831E-05 | 9.791E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 3.831E-05 | 8.812E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 4.192E-05 | 8.766E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.496E-06 | 0.01586997 |
|---|
| protein refolding | GO:0042026 |  | 8.959E-06 | 0.01094306 |
|---|
| response to UV | GO:0009411 |  | 1.117E-05 | 9.099E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.245E-05 | 7.603E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.671E-05 | 8.163E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.671E-05 | 6.802E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.671E-05 | 5.831E-03 |
|---|
| mast cell activation | GO:0045576 |  | 1.671E-05 | 5.102E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 2.215E-05 | 6.013E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.313E-05 | 5.651E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.682E-05 | 5.957E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.491E-06 | 0.01802278 |
|---|
| protein refolding | GO:0042026 |  | 9.903E-06 | 0.01191282 |
|---|
| response to UV | GO:0009411 |  | 1.152E-05 | 9.236E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.435E-05 | 8.633E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.847E-05 | 8.886E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.847E-05 | 7.405E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.847E-05 | 6.347E-03 |
|---|
| mast cell activation | GO:0045576 |  | 1.847E-05 | 5.554E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 2.337E-05 | 6.247E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.443E-05 | 5.879E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 2.901E-05 | 6.346E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.496E-06 | 0.01586997 |
|---|
| protein refolding | GO:0042026 |  | 8.959E-06 | 0.01094306 |
|---|
| response to UV | GO:0009411 |  | 1.117E-05 | 9.099E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.245E-05 | 7.603E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.671E-05 | 8.163E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.671E-05 | 6.802E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.671E-05 | 5.831E-03 |
|---|
| mast cell activation | GO:0045576 |  | 1.671E-05 | 5.102E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 2.215E-05 | 6.013E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.313E-05 | 5.651E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.682E-05 | 5.957E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.491E-06 | 0.01802278 |
|---|
| protein refolding | GO:0042026 |  | 9.903E-06 | 0.01191282 |
|---|
| response to UV | GO:0009411 |  | 1.152E-05 | 9.236E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.435E-05 | 8.633E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.847E-05 | 8.886E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.847E-05 | 7.405E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.847E-05 | 6.347E-03 |
|---|
| mast cell activation | GO:0045576 |  | 1.847E-05 | 5.554E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 2.337E-05 | 6.247E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.443E-05 | 5.879E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 2.901E-05 | 6.346E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.259E-08 | 4.494E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 3.704E-08 | 6.614E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.366E-06 | 4.006E-03 |
|---|
| response to UV | GO:0009411 |  | 4.081E-06 | 3.643E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 4.142E-06 | 2.958E-03 |
|---|
| nitric-oxide synthase regulator activity | GO:0030235 |  | 4.142E-06 | 2.465E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.339E-06 | 2.214E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.21E-06 | 2.772E-03 |
|---|
| mast cell activation | GO:0045576 |  | 6.21E-06 | 2.464E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 7.937E-06 | 2.834E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 8.338E-06 | 2.707E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 8.439E-09 | 3.078E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 5.475E-08 | 9.986E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.839E-06 | 2.236E-03 |
|---|
| response to UV | GO:0009411 |  | 2.113E-06 | 1.927E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.095E-06 | 2.258E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 3.136E-06 | 1.907E-03 |
|---|
| nitric-oxide synthase regulator activity | GO:0030235 |  | 3.136E-06 | 1.634E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.703E-06 | 2.144E-03 |
|---|
| mast cell activation | GO:0045576 |  | 4.703E-06 | 1.906E-03 |
|---|
| response to metal ion | GO:0010038 |  | 5.071E-06 | 1.85E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 6.167E-06 | 2.045E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.491E-06 | 0.01802278 |
|---|
| protein refolding | GO:0042026 |  | 9.903E-06 | 0.01191282 |
|---|
| response to UV | GO:0009411 |  | 1.152E-05 | 9.236E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.435E-05 | 8.633E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.847E-05 | 8.886E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.847E-05 | 7.405E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.847E-05 | 6.347E-03 |
|---|
| mast cell activation | GO:0045576 |  | 1.847E-05 | 5.554E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 2.337E-05 | 6.247E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.443E-05 | 5.879E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 2.901E-05 | 6.346E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.787E-05 | 0.04111132 |
|---|
| protein refolding | GO:0042026 |  | 2.056E-05 | 0.0236386 |
|---|
| response to UV | GO:0009411 |  | 2.194E-05 | 0.01681726 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 0.01653503 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 2.876E-05 | 0.01322803 |
|---|
| mast cell activation | GO:0045576 |  | 2.876E-05 | 0.01102336 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.998E-05 | 9.85E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.831E-05 | 0.01101537 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 3.831E-05 | 9.791E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 3.831E-05 | 8.812E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 4.192E-05 | 8.766E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.787E-05 | 0.04111132 |
|---|
| protein refolding | GO:0042026 |  | 2.056E-05 | 0.0236386 |
|---|
| response to UV | GO:0009411 |  | 2.194E-05 | 0.01681726 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 0.01653503 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 2.876E-05 | 0.01322803 |
|---|
| mast cell activation | GO:0045576 |  | 2.876E-05 | 0.01102336 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.998E-05 | 9.85E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.831E-05 | 0.01101537 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 3.831E-05 | 9.791E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 3.831E-05 | 8.812E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 4.192E-05 | 8.766E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.787E-05 | 0.04111132 |
|---|
| protein refolding | GO:0042026 |  | 2.056E-05 | 0.0236386 |
|---|
| response to UV | GO:0009411 |  | 2.194E-05 | 0.01681726 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 0.01653503 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 2.876E-05 | 0.01322803 |
|---|
| mast cell activation | GO:0045576 |  | 2.876E-05 | 0.01102336 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.998E-05 | 9.85E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.831E-05 | 0.01101537 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 3.831E-05 | 9.791E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 3.831E-05 | 8.812E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 4.192E-05 | 8.766E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.787E-05 | 0.04111132 |
|---|
| protein refolding | GO:0042026 |  | 2.056E-05 | 0.0236386 |
|---|
| response to UV | GO:0009411 |  | 2.194E-05 | 0.01681726 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 0.01653503 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 2.876E-05 | 0.01322803 |
|---|
| mast cell activation | GO:0045576 |  | 2.876E-05 | 0.01102336 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.998E-05 | 9.85E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.831E-05 | 0.01101537 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 3.831E-05 | 9.791E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 3.831E-05 | 8.812E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 4.192E-05 | 8.766E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein refolding | GO:0042026 |  | 2.345E-08 | 5.564E-05 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 4.1E-08 | 4.865E-05 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 6.555E-08 | 5.185E-05 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.323E-06 | 7.849E-04 |
|---|
| response to unfolded protein | GO:0006986 |  | 1.68E-06 | 7.974E-04 |
|---|
| response to protein stimulus | GO:0051789 |  | 2.143E-06 | 8.477E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.342E-06 | 7.94E-04 |
|---|
| mitochondrial membrane organization | GO:0007006 |  | 2.66E-06 | 7.89E-04 |
|---|
| short-chain fatty acid metabolic process | GO:0046459 |  | 1.166E-05 | 3.074E-03 |
|---|
| galactolipid metabolic process | GO:0019374 |  | 1.166E-05 | 2.766E-03 |
|---|
| mitochondrial transport | GO:0006839 |  | 1.409E-05 | 3.039E-03 |
|---|



