Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 581-7-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1888 | 3.169e-03 | 3.748e-03 | 4.072e-02 |
|---|
| IPC-NIBC-129 | 0.2798 | 3.622e-02 | 4.512e-02 | 3.613e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.3037 | 8.669e-03 | 1.354e-02 | 2.415e-01 |
|---|
| Loi_GPL570 | 0.2497 | 4.081e-02 | 4.049e-02 | 2.189e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1469 | 2.000e-06 | 1.300e-05 | 5.000e-06 |
|---|
| Parker_GPL1390 | 0.2816 | 1.148e-01 | 1.158e-01 | 5.773e-01 |
|---|
| Parker_GPL887 | 0.1482 | 4.907e-01 | 5.246e-01 | 5.247e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3155 | 9.479e-02 | 6.874e-02 | 5.017e-01 |
|---|
| Schmidt | 0.2643 | 1.191e-02 | 1.689e-02 | 1.495e-01 |
|---|
| Sotiriou | 0.3045 | 1.573e-03 | 4.030e-04 | 4.441e-02 |
|---|
| Wang | 0.2354 | 1.366e-01 | 9.898e-02 | 4.456e-01 |
|---|
| Zhang | 0.2764 | 4.693e-02 | 2.647e-02 | 1.197e-01 |
|---|
| Zhou | 0.4407 | 9.337e-02 | 1.025e-01 | 5.259e-01 |
|---|
Expression data for subnetwork 581-7-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
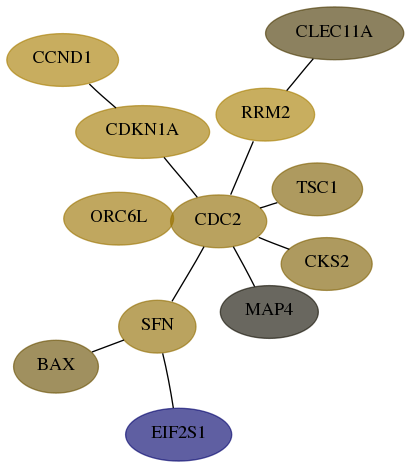
Score for each gene in subnetwork 581-7-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| rrm2 |   | 21 | 10 | 44 | 26 | 0.131 | 0.207 | 0.227 | 0.187 | 0.161 | 0.178 | 0.137 | 0.303 | 0.190 | 0.219 | 0.117 | 0.129 | 0.302 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| bax |   | 9 | 27 | 17 | 15 | 0.029 | 0.185 | 0.233 | 0.192 | 0.088 | -0.043 | 0.184 | 0.172 | 0.175 | 0.019 | -0.094 | 0.041 | 0.235 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| cks2 |   | 44 | 6 | 73 | 54 | 0.050 | 0.233 | 0.170 | 0.103 | 0.186 | 0.172 | 0.093 | 0.167 | 0.263 | 0.151 | 0.145 | 0.284 | 0.383 |
|---|
| tsc1 |   | 47 | 5 | 17 | 8 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.043 | -0.007 | 0.207 |
|---|
| orc6l |   | 8 | 33 | 73 | 61 | 0.094 | 0.270 | 0.084 | 0.114 | 0.106 | 0.240 | 0.256 | 0.234 | 0.192 | 0.105 | 0.086 | 0.175 | 0.160 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| map4 |   | 16 | 13 | 73 | 55 | 0.002 | 0.046 | 0.067 | -0.043 | 0.034 | 0.119 | 0.159 | -0.058 | -0.020 | 0.043 | 0.108 | -0.074 | 0.302 |
|---|
| sfn |   | 50 | 4 | 29 | 17 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.025 | 0.079 | 0.181 |
|---|
| eif2s1 |   | 14 | 17 | 73 | 57 | -0.031 | 0.151 | 0.253 | 0.182 | 0.098 | 0.167 | 0.005 | 0.270 | 0.233 | 0.038 | 0.122 | 0.239 | 0.265 |
|---|
| clec11a |   | 17 | 12 | 44 | 27 | 0.013 | 0.101 | 0.071 | -0.085 | 0.066 | 0.082 | 0.209 | -0.126 | -0.034 | -0.001 | 0.191 | 0.059 | -0.075 |
|---|
GO Enrichment output for subnetwork 581-7-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.085E-08 | 2.496E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 5.823E-08 | 6.697E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.018E-07 | 7.805E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.816E-06 | 2.194E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.121E-05 | 9.757E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 2.121E-05 | 8.131E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.782E-05 | 9.14E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.464E-05 | 9.96E-03 |
|---|
| response to UV | GO:0009411 |  | 4.249E-05 | 0.01085805 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.446E-05 | 0.01022621 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 4.446E-05 | 9.297E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.022E-09 | 2.496E-06 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.418E-06 | 7.839E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.771E-06 | 5.514E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.239E-06 | 4.421E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 9.622E-06 | 4.701E-03 |
|---|
| response to UV | GO:0009411 |  | 1.245E-05 | 5.069E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.346E-05 | 4.699E-03 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 1.346E-05 | 4.111E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.794E-05 | 4.87E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.794E-05 | 4.383E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 2.306E-05 | 5.121E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.789E-09 | 6.709E-06 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.113E-08 | 3.745E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 7.461E-08 | 5.984E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.356E-06 | 8.156E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 9.574E-06 | 4.607E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 9.574E-06 | 3.839E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.23E-05 | 4.229E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.315E-05 | 3.956E-03 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 2.008E-05 | 5.368E-03 |
|---|
| response to UV | GO:0009411 |  | 2.02E-05 | 4.86E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.676E-05 | 5.853E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.022E-09 | 2.496E-06 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.418E-06 | 7.839E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.771E-06 | 5.514E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.239E-06 | 4.421E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 9.622E-06 | 4.701E-03 |
|---|
| response to UV | GO:0009411 |  | 1.245E-05 | 5.069E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.346E-05 | 4.699E-03 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 1.346E-05 | 4.111E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.794E-05 | 4.87E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.794E-05 | 4.383E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 2.306E-05 | 5.121E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.789E-09 | 6.709E-06 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.113E-08 | 3.745E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 7.461E-08 | 5.984E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.356E-06 | 8.156E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 9.574E-06 | 4.607E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 9.574E-06 | 3.839E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.23E-05 | 4.229E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.315E-05 | 3.956E-03 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 2.008E-05 | 5.368E-03 |
|---|
| response to UV | GO:0009411 |  | 2.02E-05 | 4.86E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.676E-05 | 5.853E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 5.533E-11 | 1.976E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.667E-10 | 2.977E-07 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 5.971E-10 | 7.107E-07 |
|---|
| kinase regulator activity | GO:0019207 |  | 1.54E-09 | 1.375E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.73E-08 | 1.236E-05 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 1.122E-07 | 6.677E-05 |
|---|
| protein kinase inhibitor activity | GO:0004860 |  | 1.004E-06 | 5.121E-04 |
|---|
| kinase inhibitor activity | GO:0019210 |  | 1.243E-06 | 5.549E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.759E-06 | 1.491E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.618E-06 | 1.649E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.061E-06 | 1.643E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.578E-11 | 3.494E-07 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 9.653E-11 | 1.761E-07 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 3.627E-10 | 4.41E-07 |
|---|
| kinase regulator activity | GO:0019207 |  | 7.474E-10 | 6.816E-07 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 9.676E-09 | 7.06E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.125E-08 | 6.84E-06 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 7.296E-08 | 3.802E-05 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 1.122E-07 | 5.115E-05 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 2.659E-07 | 1.078E-04 |
|---|
| protein kinase inhibitor activity | GO:0004860 |  | 1.191E-06 | 4.344E-04 |
|---|
| kinase inhibitor activity | GO:0019210 |  | 1.302E-06 | 4.318E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.789E-09 | 6.709E-06 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.113E-08 | 3.745E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 7.461E-08 | 5.984E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.356E-06 | 8.156E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 9.574E-06 | 4.607E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 9.574E-06 | 3.839E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.23E-05 | 4.229E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.315E-05 | 3.956E-03 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 2.008E-05 | 5.368E-03 |
|---|
| response to UV | GO:0009411 |  | 2.02E-05 | 4.86E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.676E-05 | 5.853E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.085E-08 | 2.496E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 5.823E-08 | 6.697E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.018E-07 | 7.805E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.816E-06 | 2.194E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.121E-05 | 9.757E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 2.121E-05 | 8.131E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.782E-05 | 9.14E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.464E-05 | 9.96E-03 |
|---|
| response to UV | GO:0009411 |  | 4.249E-05 | 0.01085805 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.446E-05 | 0.01022621 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 4.446E-05 | 9.297E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.085E-08 | 2.496E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 5.823E-08 | 6.697E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.018E-07 | 7.805E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.816E-06 | 2.194E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.121E-05 | 9.757E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 2.121E-05 | 8.131E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.782E-05 | 9.14E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.464E-05 | 9.96E-03 |
|---|
| response to UV | GO:0009411 |  | 4.249E-05 | 0.01085805 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.446E-05 | 0.01022621 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 4.446E-05 | 9.297E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.085E-08 | 2.496E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 5.823E-08 | 6.697E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.018E-07 | 7.805E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.816E-06 | 2.194E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.121E-05 | 9.757E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 2.121E-05 | 8.131E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.782E-05 | 9.14E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.464E-05 | 9.96E-03 |
|---|
| response to UV | GO:0009411 |  | 4.249E-05 | 0.01085805 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.446E-05 | 0.01022621 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 4.446E-05 | 9.297E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.085E-08 | 2.496E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 5.823E-08 | 6.697E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.018E-07 | 7.805E-05 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.816E-06 | 2.194E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.121E-05 | 9.757E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 2.121E-05 | 8.131E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.782E-05 | 9.14E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.464E-05 | 9.96E-03 |
|---|
| response to UV | GO:0009411 |  | 4.249E-05 | 0.01085805 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.446E-05 | 0.01022621 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 4.446E-05 | 9.297E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.039E-08 | 2.466E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 8.714E-08 | 1.034E-04 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.393E-07 | 1.102E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.78E-06 | 2.243E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.906E-05 | 9.048E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.765E-05 | 0.01093716 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 2.857E-05 | 9.686E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.184E-05 | 9.445E-03 |
|---|
| response to UV | GO:0009411 |  | 3.888E-05 | 0.01025042 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.997E-05 | 9.484E-03 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 3.997E-05 | 8.622E-03 |
|---|



