Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 58-7-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1686 | 3.330e-03 | 3.940e-03 | 4.235e-02 |
|---|
| IPC-NIBC-129 | 0.2655 | 2.204e-02 | 2.641e-02 | 2.450e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.3014 | 1.965e-02 | 2.827e-02 | 3.856e-01 |
|---|
| Loi_GPL570 | 0.2112 | 4.974e-02 | 4.937e-02 | 2.539e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1670 | 1.000e-06 | 9.000e-06 | 2.000e-06 |
|---|
| Parker_GPL1390 | 0.3172 | 1.515e-01 | 1.562e-01 | 6.530e-01 |
|---|
| Parker_GPL887 | 0.2166 | 3.202e-01 | 3.545e-01 | 3.272e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3745 | 1.268e-01 | 9.628e-02 | 5.946e-01 |
|---|
| Schmidt | 0.2712 | 2.472e-03 | 3.739e-03 | 4.151e-02 |
|---|
| Sotiriou | 0.3998 | 2.049e-03 | 5.630e-04 | 5.394e-02 |
|---|
| Wang | 0.1568 | 9.891e-02 | 6.789e-02 | 3.629e-01 |
|---|
| Zhang | 0.2480 | 1.458e-01 | 1.028e-01 | 3.327e-01 |
|---|
| Zhou | 0.4248 | 5.337e-02 | 5.570e-02 | 3.362e-01 |
|---|
Expression data for subnetwork 58-7-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
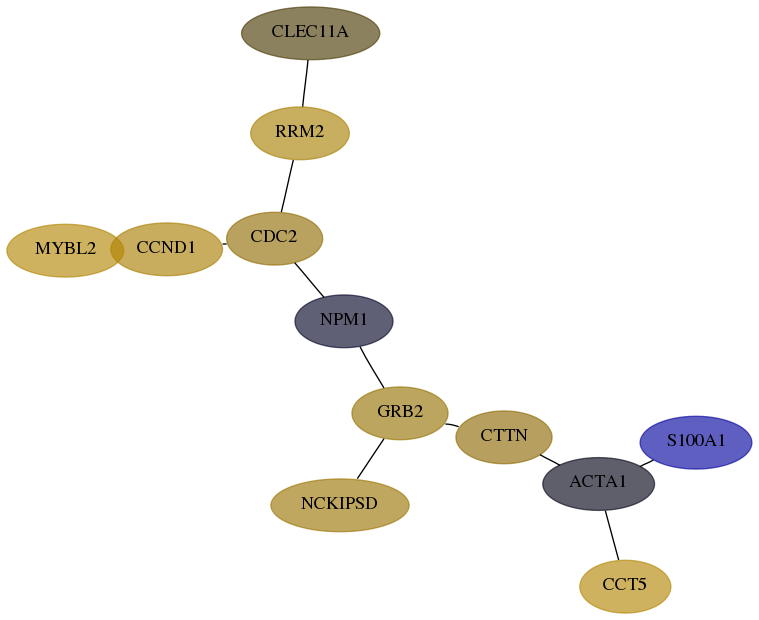
Score for each gene in subnetwork 58-7-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| rrm2 |   | 21 | 10 | 44 | 26 | 0.131 | 0.207 | 0.227 | 0.187 | 0.161 | 0.178 | 0.137 | 0.303 | 0.190 | 0.219 | 0.117 | 0.129 | 0.302 |
|---|
| mybl2 |   | 3 | 79 | 44 | 44 | 0.162 | 0.237 | 0.166 | 0.079 | 0.124 | 0.184 | 0.333 | 0.275 | 0.202 | 0.182 | 0.120 | 0.074 | 0.245 |
|---|
| cttn |   | 9 | 27 | 44 | 37 | 0.067 | 0.065 | 0.123 | 0.238 | 0.126 | 0.263 | -0.172 | 0.216 | 0.069 | 0.115 | 0.129 | 0.025 | 0.279 |
|---|
| s100a1 |   | 2 | 104 | 44 | 52 | -0.100 | 0.147 | 0.022 | -0.057 | 0.048 | 0.011 | 0.134 | 0.085 | 0.103 | 0.032 | 0.027 | 0.014 | -0.049 |
|---|
| cct5 |   | 2 | 104 | 44 | 52 | 0.165 | 0.258 | 0.182 | 0.155 | 0.209 | 0.212 | 0.257 | 0.353 | 0.143 | 0.204 | 0.106 | 0.039 | 0.289 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| nckipsd |   | 3 | 79 | 44 | 44 | 0.094 | 0.086 | 0.221 | -0.049 | 0.109 | -0.007 | 0.119 | 0.121 | 0.006 | 0.126 | 0.073 | 0.054 | 0.339 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| acta1 |   | 6 | 43 | 44 | 40 | -0.003 | 0.098 | -0.008 | -0.030 | 0.119 | 0.174 | 0.376 | 0.024 | 0.182 | 0.214 | -0.013 | 0.048 | 0.201 |
|---|
| clec11a |   | 17 | 12 | 44 | 27 | 0.013 | 0.101 | 0.071 | -0.085 | 0.066 | 0.082 | 0.209 | -0.126 | -0.034 | -0.001 | 0.191 | 0.059 | -0.075 |
|---|
| npm1 |   | 4 | 60 | 44 | 41 | -0.005 | 0.172 | 0.085 | 0.197 | 0.093 | 0.221 | -0.227 | -0.050 | 0.065 | 0.169 | -0.000 | -0.086 | 0.199 |
|---|
| grb2 |   | 9 | 27 | 1 | 4 | 0.084 | -0.013 | 0.086 | -0.031 | 0.162 | -0.038 | 0.003 | 0.065 | 0.158 | 0.127 | -0.097 | 0.194 | -0.005 |
|---|
GO Enrichment output for subnetwork 58-7-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.364E-11 | 3.138E-08 |
|---|
| ribosome assembly | GO:0042255 |  | 8.956E-10 | 1.03E-06 |
|---|
| centrosome cycle | GO:0007098 |  | 3.691E-09 | 2.83E-06 |
|---|
| cell aging | GO:0007569 |  | 1.609E-08 | 9.254E-06 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 1.965E-08 | 9.04E-06 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 4.002E-08 | 1.534E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.534E-08 | 1.49E-05 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 7.309E-08 | 2.101E-05 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 7.309E-08 | 1.868E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 7.928E-08 | 1.823E-05 |
|---|
| positive regulation of transcription factor activity | GO:0051091 |  | 1.562E-07 | 3.267E-05 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.319E-12 | 3.222E-09 |
|---|
| ribosome assembly | GO:0042255 |  | 2.628E-10 | 3.211E-07 |
|---|
| centrosome cycle | GO:0007098 |  | 8.018E-10 | 6.529E-07 |
|---|
| cell aging | GO:0007569 |  | 1.913E-09 | 1.168E-06 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 3.901E-09 | 1.906E-06 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 8.189E-09 | 3.334E-06 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 9.354E-09 | 3.265E-06 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 1.714E-08 | 5.233E-06 |
|---|
| positive regulation of transcription factor activity | GO:0051091 |  | 3.192E-08 | 8.665E-06 |
|---|
| positive regulation of DNA binding | GO:0043388 |  | 5.02E-08 | 1.226E-05 |
|---|
| positive regulation of binding | GO:0051099 |  | 6.97E-08 | 1.548E-05 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 2.559E-12 | 6.156E-09 |
|---|
| ribosome assembly | GO:0042255 |  | 3.641E-10 | 4.38E-07 |
|---|
| centrosome cycle | GO:0007098 |  | 1.554E-09 | 1.246E-06 |
|---|
| cell aging | GO:0007569 |  | 3.033E-09 | 1.824E-06 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 7.553E-09 | 3.635E-06 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 1.198E-08 | 4.803E-06 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 1.81E-08 | 6.222E-06 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.243E-08 | 6.747E-06 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 2.631E-08 | 7.033E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 5.378E-08 | 1.294E-05 |
|---|
| positive regulation of transcription factor activity | GO:0051091 |  | 5.6E-08 | 1.225E-05 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.319E-12 | 3.222E-09 |
|---|
| ribosome assembly | GO:0042255 |  | 2.628E-10 | 3.211E-07 |
|---|
| centrosome cycle | GO:0007098 |  | 8.018E-10 | 6.529E-07 |
|---|
| cell aging | GO:0007569 |  | 1.913E-09 | 1.168E-06 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 3.901E-09 | 1.906E-06 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 8.189E-09 | 3.334E-06 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 9.354E-09 | 3.265E-06 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 1.714E-08 | 5.233E-06 |
|---|
| positive regulation of transcription factor activity | GO:0051091 |  | 3.192E-08 | 8.665E-06 |
|---|
| positive regulation of DNA binding | GO:0043388 |  | 5.02E-08 | 1.226E-05 |
|---|
| positive regulation of binding | GO:0051099 |  | 6.97E-08 | 1.548E-05 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 2.559E-12 | 6.156E-09 |
|---|
| ribosome assembly | GO:0042255 |  | 3.641E-10 | 4.38E-07 |
|---|
| centrosome cycle | GO:0007098 |  | 1.554E-09 | 1.246E-06 |
|---|
| cell aging | GO:0007569 |  | 3.033E-09 | 1.824E-06 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 7.553E-09 | 3.635E-06 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 1.198E-08 | 4.803E-06 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 1.81E-08 | 6.222E-06 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.243E-08 | 6.747E-06 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 2.631E-08 | 7.033E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 5.378E-08 | 1.294E-05 |
|---|
| positive regulation of transcription factor activity | GO:0051091 |  | 5.6E-08 | 1.225E-05 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| skeletal myofibril assembly | GO:0014866 |  | 5.061E-06 | 0.0180734 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.589E-06 | 0.01354953 |
|---|
| insulin receptor substrate binding | GO:0043560 |  | 7.589E-06 | 9.033E-03 |
|---|
| oxidoreductase activity. acting on CH or CH2 groups | GO:0016725 |  | 1.062E-05 | 9.481E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.062E-05 | 7.585E-03 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.415E-05 | 8.424E-03 |
|---|
| epidermal growth factor receptor binding | GO:0005154 |  | 1.415E-05 | 7.221E-03 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.819E-05 | 8.12E-03 |
|---|
| ribosome assembly | GO:0042255 |  | 2.273E-05 | 9.018E-03 |
|---|
| ADP binding | GO:0043531 |  | 2.777E-05 | 9.916E-03 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 2.777E-05 | 9.015E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| muscle thin filament assembly | GO:0030240 |  | 5.187E-06 | 0.0189226 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 5.187E-06 | 9.461E-03 |
|---|
| epidermal growth factor receptor binding | GO:0005154 |  | 5.187E-06 | 6.308E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.777E-06 | 7.093E-03 |
|---|
| skeletal myofibril assembly | GO:0014866 |  | 7.777E-06 | 5.674E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 7.777E-06 | 4.729E-03 |
|---|
| insulin receptor substrate binding | GO:0043560 |  | 1.088E-05 | 5.672E-03 |
|---|
| oxidoreductase activity. acting on CH or CH2 groups | GO:0016725 |  | 1.088E-05 | 4.963E-03 |
|---|
| ATPase binding | GO:0051117 |  | 1.088E-05 | 4.411E-03 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.451E-05 | 5.292E-03 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.451E-05 | 4.81E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 2.559E-12 | 6.156E-09 |
|---|
| ribosome assembly | GO:0042255 |  | 3.641E-10 | 4.38E-07 |
|---|
| centrosome cycle | GO:0007098 |  | 1.554E-09 | 1.246E-06 |
|---|
| cell aging | GO:0007569 |  | 3.033E-09 | 1.824E-06 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 7.553E-09 | 3.635E-06 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 1.198E-08 | 4.803E-06 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 1.81E-08 | 6.222E-06 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.243E-08 | 6.747E-06 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 2.631E-08 | 7.033E-06 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 5.378E-08 | 1.294E-05 |
|---|
| positive regulation of transcription factor activity | GO:0051091 |  | 5.6E-08 | 1.225E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.364E-11 | 3.138E-08 |
|---|
| ribosome assembly | GO:0042255 |  | 8.956E-10 | 1.03E-06 |
|---|
| centrosome cycle | GO:0007098 |  | 3.691E-09 | 2.83E-06 |
|---|
| cell aging | GO:0007569 |  | 1.609E-08 | 9.254E-06 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 1.965E-08 | 9.04E-06 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 4.002E-08 | 1.534E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.534E-08 | 1.49E-05 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 7.309E-08 | 2.101E-05 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 7.309E-08 | 1.868E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 7.928E-08 | 1.823E-05 |
|---|
| positive regulation of transcription factor activity | GO:0051091 |  | 1.562E-07 | 3.267E-05 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.364E-11 | 3.138E-08 |
|---|
| ribosome assembly | GO:0042255 |  | 8.956E-10 | 1.03E-06 |
|---|
| centrosome cycle | GO:0007098 |  | 3.691E-09 | 2.83E-06 |
|---|
| cell aging | GO:0007569 |  | 1.609E-08 | 9.254E-06 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 1.965E-08 | 9.04E-06 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 4.002E-08 | 1.534E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.534E-08 | 1.49E-05 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 7.309E-08 | 2.101E-05 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 7.309E-08 | 1.868E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 7.928E-08 | 1.823E-05 |
|---|
| positive regulation of transcription factor activity | GO:0051091 |  | 1.562E-07 | 3.267E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.364E-11 | 3.138E-08 |
|---|
| ribosome assembly | GO:0042255 |  | 8.956E-10 | 1.03E-06 |
|---|
| centrosome cycle | GO:0007098 |  | 3.691E-09 | 2.83E-06 |
|---|
| cell aging | GO:0007569 |  | 1.609E-08 | 9.254E-06 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 1.965E-08 | 9.04E-06 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 4.002E-08 | 1.534E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.534E-08 | 1.49E-05 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 7.309E-08 | 2.101E-05 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 7.309E-08 | 1.868E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 7.928E-08 | 1.823E-05 |
|---|
| positive regulation of transcription factor activity | GO:0051091 |  | 1.562E-07 | 3.267E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.364E-11 | 3.138E-08 |
|---|
| ribosome assembly | GO:0042255 |  | 8.956E-10 | 1.03E-06 |
|---|
| centrosome cycle | GO:0007098 |  | 3.691E-09 | 2.83E-06 |
|---|
| cell aging | GO:0007569 |  | 1.609E-08 | 9.254E-06 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 1.965E-08 | 9.04E-06 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 4.002E-08 | 1.534E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.534E-08 | 1.49E-05 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 7.309E-08 | 2.101E-05 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 7.309E-08 | 1.868E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 7.928E-08 | 1.823E-05 |
|---|
| positive regulation of transcription factor activity | GO:0051091 |  | 1.562E-07 | 3.267E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of centrosome duplication | GO:0010824 |  | 8.146E-12 | 1.933E-08 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 2.442E-11 | 2.897E-08 |
|---|
| ribosome assembly | GO:0042255 |  | 2.206E-09 | 1.745E-06 |
|---|
| centrosome cycle | GO:0007098 |  | 2.939E-09 | 1.744E-06 |
|---|
| microtubule organizing center organization | GO:0031023 |  | 1.422E-08 | 6.749E-06 |
|---|
| cell aging | GO:0007569 |  | 1.705E-08 | 6.743E-06 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 3.796E-08 | 1.287E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 5.387E-08 | 1.598E-05 |
|---|
| positive regulation of NF-kappaB transcription factor activity | GO:0051092 |  | 5.733E-08 | 1.512E-05 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 6.518E-08 | 1.547E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 8.612E-08 | 1.858E-05 |
|---|



