Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 5714-7-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2110 | 5.092e-03 | 6.053e-03 | 5.916e-02 |
|---|
| IPC-NIBC-129 | 0.2265 | 3.023e-02 | 3.716e-02 | 3.154e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2809 | 4.338e-03 | 7.264e-03 | 1.552e-01 |
|---|
| Loi_GPL570 | 0.2819 | 8.453e-02 | 8.400e-02 | 3.698e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1386 | 9.000e-06 | 6.100e-05 | 6.800e-05 |
|---|
| Parker_GPL1390 | 0.2946 | 8.423e-02 | 8.270e-02 | 4.970e-01 |
|---|
| Parker_GPL887 | 0.4689 | 5.794e-02 | 7.307e-02 | 4.303e-02 |
|---|
| Pawitan_GPL96-GPL97 | 0.3959 | 9.207e-01 | 9.219e-01 | 1.000e+00 |
|---|
| Schmidt | 0.2402 | 6.360e-03 | 9.263e-03 | 9.145e-02 |
|---|
| Sotiriou | 0.3427 | 5.669e-03 | 2.032e-03 | 1.110e-01 |
|---|
| Wang | 0.2343 | 1.557e-01 | 1.153e-01 | 4.826e-01 |
|---|
| Zhang | 0.0228 | 4.770e-02 | 2.699e-02 | 1.216e-01 |
|---|
| Zhou | 0.3634 | 4.347e-02 | 4.443e-02 | 2.787e-01 |
|---|
Expression data for subnetwork 5714-7-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
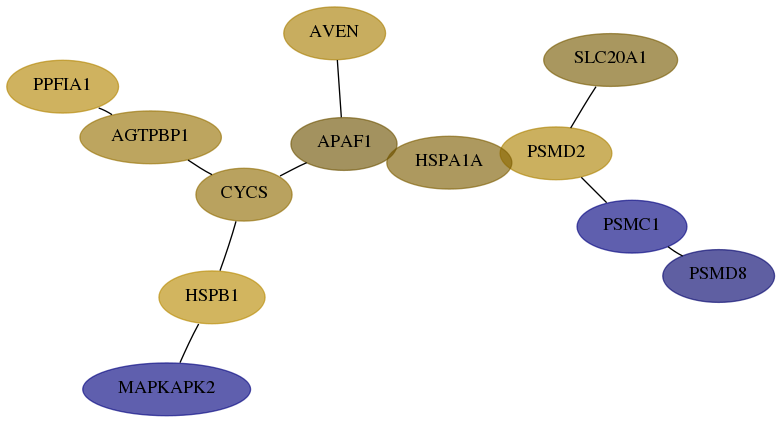
Score for each gene in subnetwork 5714-7-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| agtpbp1 |   | 11 | 21 | 17 | 13 | 0.086 | 0.219 | 0.009 | 0.181 | 0.071 | 0.078 | 0.296 | 0.026 | 0.110 | 0.151 | 0.103 | 0.064 | 0.226 |
|---|
| psmd2 |   | 7 | 38 | 37 | 28 | 0.147 | 0.322 | 0.122 | 0.146 | 0.143 | 0.246 | 0.444 | 0.194 | 0.318 | 0.151 | 0.154 | 0.042 | 0.207 |
|---|
| cycs |   | 10 | 25 | 17 | 14 | 0.073 | 0.206 | 0.080 | 0.094 | 0.117 | -0.056 | 0.202 | 0.272 | 0.157 | 0.077 | 0.190 | 0.135 | 0.234 |
|---|
| ppfia1 |   | 8 | 33 | 17 | 16 | 0.164 | 0.080 | 0.149 | 0.269 | 0.151 | 0.214 | 0.093 | 0.225 | 0.036 | 0.176 | 0.153 | 0.127 | 0.272 |
|---|
| psmd8 |   | 1 | 164 | 61 | 84 | -0.031 | 0.158 | 0.163 | 0.198 | 0.168 | 0.091 | -0.155 | 0.274 | -0.009 | 0.177 | 0.086 | -0.069 | 0.172 |
|---|
| hspb1 |   | 19 | 11 | 17 | 11 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | -0.026 | -0.065 | 0.289 | -0.021 | 0.201 | 0.125 | -0.077 | 0.290 |
|---|
| psmc1 |   | 2 | 104 | 61 | 69 | -0.048 | 0.159 | 0.166 | 0.140 | 0.109 | 0.032 | 0.072 | 0.250 | 0.161 | 0.120 | 0.086 | 0.214 | 0.275 |
|---|
| mapkapk2 |   | 13 | 19 | 17 | 12 | -0.050 | 0.102 | 0.156 | -0.116 | 0.090 | 0.168 | 0.303 | 0.074 | 0.171 | 0.117 | 0.085 | -0.124 | 0.312 |
|---|
| aven |   | 1 | 164 | 61 | 84 | 0.130 | 0.211 | 0.177 | 0.009 | 0.093 | 0.127 | 0.194 | 0.196 | 0.071 | 0.094 | 0.081 | -0.165 | 0.220 |
|---|
| slc20a1 |   | 7 | 38 | 37 | 28 | 0.041 | 0.039 | 0.109 | 0.170 | 0.101 | 0.121 | 0.326 | 0.086 | 0.048 | 0.124 | 0.121 | -0.050 | 0.299 |
|---|
| apaf1 |   | 2 | 104 | 37 | 46 | 0.033 | 0.135 | 0.021 | -0.049 | 0.072 | 0.087 | 0.202 | 0.001 | 0.120 | 0.008 | 0.026 | -0.004 | 0.228 |
|---|
| hspa1a |   | 4 | 60 | 37 | 39 | 0.047 | -0.046 | 0.070 | 0.019 | 0.160 | 0.104 | 0.230 | 0.108 | 0.121 | 0.110 | 0.175 | -0.055 | 0.192 |
|---|
GO Enrichment output for subnetwork 5714-7-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.555E-08 | 5.877E-05 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 5.829E-07 | 6.703E-04 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 6.233E-07 | 4.779E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 6.233E-07 | 3.584E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 6.658E-07 | 3.063E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 8.063E-07 | 3.091E-04 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 8.063E-07 | 2.649E-04 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 1.027E-06 | 2.951E-04 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.152E-06 | 2.944E-04 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.437E-06 | 3.305E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.863E-06 | 3.895E-04 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.329E-11 | 3.247E-08 |
|---|
| activation of caspase activity | GO:0006919 |  | 4.287E-08 | 5.237E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 7.329E-08 | 5.968E-05 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 1.415E-07 | 8.644E-05 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 1.503E-07 | 7.344E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.503E-07 | 6.12E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.595E-07 | 5.566E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.895E-07 | 5.788E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 1.895E-07 | 5.145E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.076E-07 | 5.073E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 2.118E-07 | 4.703E-05 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.166E-11 | 5.211E-08 |
|---|
| activation of caspase activity | GO:0006919 |  | 5.893E-08 | 7.09E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.029E-07 | 8.254E-05 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 2.298E-07 | 1.382E-04 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 2.44E-07 | 1.174E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.44E-07 | 9.786E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 2.589E-07 | 8.9E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 3.001E-07 | 9.026E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 3.076E-07 | 8.224E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 3.076E-07 | 7.402E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 3.076E-07 | 6.729E-05 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.329E-11 | 3.247E-08 |
|---|
| activation of caspase activity | GO:0006919 |  | 4.287E-08 | 5.237E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 7.329E-08 | 5.968E-05 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 1.415E-07 | 8.644E-05 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 1.503E-07 | 7.344E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.503E-07 | 6.12E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.595E-07 | 5.566E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.895E-07 | 5.788E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 1.895E-07 | 5.145E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.076E-07 | 5.073E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 2.118E-07 | 4.703E-05 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.166E-11 | 5.211E-08 |
|---|
| activation of caspase activity | GO:0006919 |  | 5.893E-08 | 7.09E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.029E-07 | 8.254E-05 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 2.298E-07 | 1.382E-04 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 2.44E-07 | 1.174E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.44E-07 | 9.786E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 2.589E-07 | 8.9E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 3.001E-07 | 9.026E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 3.076E-07 | 8.224E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 3.076E-07 | 7.402E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 3.076E-07 | 6.729E-05 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| proteasome regulatory particle | GO:0005838 |  | 1.442E-08 | 5.149E-05 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.442E-08 | 2.575E-05 |
|---|
| proteasome complex | GO:0000502 |  | 4.415E-08 | 5.255E-05 |
|---|
| proteasome accessory complex | GO:0022624 |  | 4.937E-08 | 4.408E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.284E-07 | 9.169E-05 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 1.372E-07 | 8.164E-05 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 1.464E-07 | 7.469E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.561E-07 | 6.968E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.663E-07 | 6.597E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 1.881E-07 | 6.716E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 2.119E-07 | 6.88E-05 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.454E-08 | 5.304E-05 |
|---|
| proteasome regulatory particle | GO:0005838 |  | 2.325E-08 | 4.241E-05 |
|---|
| proteasome complex | GO:0000502 |  | 5.086E-08 | 6.184E-05 |
|---|
| proteasome accessory complex | GO:0022624 |  | 6.841E-08 | 6.239E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.449E-07 | 1.057E-04 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 1.449E-07 | 8.807E-05 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 1.648E-07 | 8.59E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.756E-07 | 8.005E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.986E-07 | 8.048E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 2.109E-07 | 7.692E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 2.372E-07 | 7.866E-05 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.166E-11 | 5.211E-08 |
|---|
| activation of caspase activity | GO:0006919 |  | 5.893E-08 | 7.09E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.029E-07 | 8.254E-05 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 2.298E-07 | 1.382E-04 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 2.44E-07 | 1.174E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.44E-07 | 9.786E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 2.589E-07 | 8.9E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 3.001E-07 | 9.026E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 3.076E-07 | 8.224E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 3.076E-07 | 7.402E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 3.076E-07 | 6.729E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.555E-08 | 5.877E-05 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 5.829E-07 | 6.703E-04 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 6.233E-07 | 4.779E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 6.233E-07 | 3.584E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 6.658E-07 | 3.063E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 8.063E-07 | 3.091E-04 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 8.063E-07 | 2.649E-04 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 1.027E-06 | 2.951E-04 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.152E-06 | 2.944E-04 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.437E-06 | 3.305E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.863E-06 | 3.895E-04 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.555E-08 | 5.877E-05 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 5.829E-07 | 6.703E-04 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 6.233E-07 | 4.779E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 6.233E-07 | 3.584E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 6.658E-07 | 3.063E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 8.063E-07 | 3.091E-04 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 8.063E-07 | 2.649E-04 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 1.027E-06 | 2.951E-04 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.152E-06 | 2.944E-04 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.437E-06 | 3.305E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.863E-06 | 3.895E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.555E-08 | 5.877E-05 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 5.829E-07 | 6.703E-04 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 6.233E-07 | 4.779E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 6.233E-07 | 3.584E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 6.658E-07 | 3.063E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 8.063E-07 | 3.091E-04 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 8.063E-07 | 2.649E-04 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 1.027E-06 | 2.951E-04 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.152E-06 | 2.944E-04 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.437E-06 | 3.305E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.863E-06 | 3.895E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.555E-08 | 5.877E-05 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 5.829E-07 | 6.703E-04 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 6.233E-07 | 4.779E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 6.233E-07 | 3.584E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 6.658E-07 | 3.063E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 8.063E-07 | 3.091E-04 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 8.063E-07 | 2.649E-04 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 1.027E-06 | 2.951E-04 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.152E-06 | 2.944E-04 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.437E-06 | 3.305E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.863E-06 | 3.895E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.476E-08 | 3.502E-05 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 4.129E-07 | 4.899E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 4.129E-07 | 3.266E-04 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 4.389E-07 | 2.604E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 5.24E-07 | 2.487E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 6.21E-07 | 2.456E-04 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 6.21E-07 | 2.105E-04 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 7.306E-07 | 2.167E-04 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 8.541E-07 | 2.252E-04 |
|---|
| regulation of ligase activity | GO:0051340 |  | 9.924E-07 | 2.355E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.147E-06 | 2.474E-04 |
|---|



