Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 55781-6-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2036 | 9.166e-03 | 1.095e-02 | 9.281e-02 |
|---|
| IPC-NIBC-129 | 0.2370 | 6.618e-02 | 8.563e-02 | 5.394e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2525 | 2.390e-02 | 3.371e-02 | 4.268e-01 |
|---|
| Loi_GPL570 | 0.2019 | 7.339e-02 | 7.290e-02 | 3.356e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1762 | 1.600e-05 | 1.010e-04 | 1.610e-04 |
|---|
| Parker_GPL1390 | 0.2378 | 9.349e-02 | 9.264e-02 | 5.234e-01 |
|---|
| Parker_GPL887 | 0.0708 | 7.589e-01 | 7.808e-01 | 8.148e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3628 | 1.459e-01 | 1.133e-01 | 6.411e-01 |
|---|
| Schmidt | 0.2320 | 1.350e-02 | 1.904e-02 | 1.643e-01 |
|---|
| Sotiriou | 0.2968 | 1.053e-03 | 2.420e-04 | 3.290e-02 |
|---|
| Wang | 0.1501 | 8.535e-02 | 5.715e-02 | 3.292e-01 |
|---|
| Zhang | 0.2341 | 1.601e-01 | 1.150e-01 | 3.598e-01 |
|---|
| Zhou | 0.4647 | 5.969e-02 | 6.297e-02 | 3.705e-01 |
|---|
Expression data for subnetwork 55781-6-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
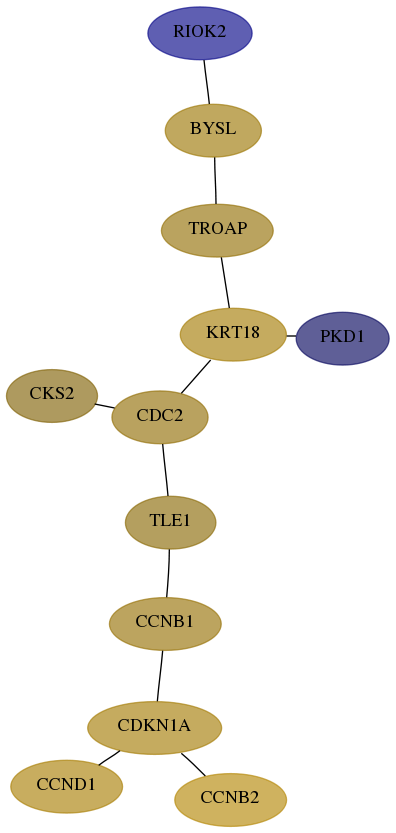
Score for each gene in subnetwork 55781-6-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| ccnb2 |   | 16 | 13 | 153 | 125 | 0.164 | 0.176 | 0.204 | 0.123 | 0.172 | 0.178 | 0.090 | 0.366 | 0.240 | 0.169 | 0.144 | 0.211 | 0.463 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| tle1 |   | 23 | 9 | 78 | 60 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | -0.028 | 0.068 | 0.025 |
|---|
| krt18 |   | 15 | 15 | 230 | 195 | 0.112 | -0.078 | 0.147 | 0.150 | 0.161 | -0.086 | -0.204 | 0.083 | -0.099 | 0.118 | 0.086 | -0.114 | 0.222 |
|---|
| ccnb1 |   | 36 | 7 | 78 | 59 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | 0.152 | 0.167 | 0.398 |
|---|
| bysl |   | 2 | 104 | 230 | 224 | 0.097 | 0.204 | 0.042 | 0.257 | 0.107 | 0.153 | 0.042 | 0.316 | 0.233 | 0.106 | -0.015 | -0.010 | 0.286 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| cks2 |   | 44 | 6 | 73 | 54 | 0.050 | 0.233 | 0.170 | 0.103 | 0.186 | 0.172 | 0.093 | 0.167 | 0.263 | 0.151 | 0.145 | 0.284 | 0.383 |
|---|
| riok2 |   | 1 | 164 | 352 | 352 | -0.058 | 0.116 | 0.025 | -0.040 | 0.096 | 0.070 | -0.000 | 0.098 | -0.012 | 0.102 | 0.097 | 0.011 | 0.199 |
|---|
| pkd1 |   | 9 | 27 | 234 | 205 | -0.021 | 0.055 | 0.007 | 0.037 | 0.071 | 0.184 | 0.256 | -0.217 | -0.068 | -0.001 | 0.046 | 0.077 | 0.151 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| troap |   | 2 | 104 | 230 | 224 | 0.078 | 0.179 | 0.137 | 0.149 | 0.119 | 0.178 | 0.538 | 0.144 | 0.284 | 0.117 | 0.050 | 0.242 | 0.488 |
|---|
GO Enrichment output for subnetwork 55781-6-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.411E-07 | 5.544E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.43E-06 | 1.644E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2E-06 | 1.533E-03 |
|---|
| interphase | GO:0051325 |  | 2.193E-06 | 1.261E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.307E-05 | 6.011E-03 |
|---|
| response to UV | GO:0009411 |  | 1.604E-05 | 6.149E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 7.697E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.121E-05 | 8.974E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 3.121E-05 | 7.977E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.764E-05 | 8.657E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.953E-05 | 8.265E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.142E-08 | 1.256E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.621E-07 | 5.644E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 5.393E-07 | 4.391E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.528E-06 | 2.155E-03 |
|---|
| response to UV | GO:0009411 |  | 6.074E-06 | 2.968E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 8.394E-06 | 3.418E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 8.394E-06 | 2.93E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.119E-05 | 3.417E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.154E-05 | 3.132E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.206E-05 | 2.946E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.371E-05 | 3.045E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.735E-08 | 1.62E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.335E-07 | 6.418E-04 |
|---|
| interphase | GO:0051325 |  | 6.046E-07 | 4.849E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 6.654E-07 | 4.002E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.35E-06 | 2.093E-03 |
|---|
| response to UV | GO:0009411 |  | 6.692E-06 | 2.684E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 9.704E-06 | 3.335E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 9.704E-06 | 2.918E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.293E-05 | 3.457E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.422E-05 | 3.422E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.486E-05 | 3.25E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.142E-08 | 1.256E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.621E-07 | 5.644E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 5.393E-07 | 4.391E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.528E-06 | 2.155E-03 |
|---|
| response to UV | GO:0009411 |  | 6.074E-06 | 2.968E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 8.394E-06 | 3.418E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 8.394E-06 | 2.93E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.119E-05 | 3.417E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.154E-05 | 3.132E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.206E-05 | 2.946E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.371E-05 | 3.045E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.735E-08 | 1.62E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.335E-07 | 6.418E-04 |
|---|
| interphase | GO:0051325 |  | 6.046E-07 | 4.849E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 6.654E-07 | 4.002E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.35E-06 | 2.093E-03 |
|---|
| response to UV | GO:0009411 |  | 6.692E-06 | 2.684E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 9.704E-06 | 3.335E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 9.704E-06 | 2.918E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.293E-05 | 3.457E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.422E-05 | 3.422E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.486E-05 | 3.25E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 5.533E-11 | 1.976E-07 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.73E-08 | 3.089E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.805E-08 | 4.529E-05 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 1.044E-07 | 9.321E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.093E-07 | 1.495E-04 |
|---|
| kinase regulator activity | GO:0019207 |  | 2.213E-07 | 1.317E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.854E-07 | 1.456E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.618E-06 | 2.061E-03 |
|---|
| response to UV | GO:0009411 |  | 5.598E-06 | 2.221E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.589E-06 | 2.71E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 7.589E-06 | 2.464E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.163E-09 | 4.242E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 6.789E-08 | 1.238E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.826E-07 | 3.436E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 6.749E-07 | 6.156E-04 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 8.061E-07 | 5.881E-04 |
|---|
| kinase regulator activity | GO:0019207 |  | 1.425E-06 | 8.661E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.841E-06 | 9.592E-04 |
|---|
| interphase | GO:0051325 |  | 2.03E-06 | 9.257E-04 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 1.196E-05 | 4.848E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 1.196E-05 | 4.363E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.459E-05 | 4.838E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.735E-08 | 1.62E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.335E-07 | 6.418E-04 |
|---|
| interphase | GO:0051325 |  | 6.046E-07 | 4.849E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 6.654E-07 | 4.002E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.35E-06 | 2.093E-03 |
|---|
| response to UV | GO:0009411 |  | 6.692E-06 | 2.684E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 9.704E-06 | 3.335E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 9.704E-06 | 2.918E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.293E-05 | 3.457E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.422E-05 | 3.422E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.486E-05 | 3.25E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.411E-07 | 5.544E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.43E-06 | 1.644E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2E-06 | 1.533E-03 |
|---|
| interphase | GO:0051325 |  | 2.193E-06 | 1.261E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.307E-05 | 6.011E-03 |
|---|
| response to UV | GO:0009411 |  | 1.604E-05 | 6.149E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 7.697E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.121E-05 | 8.974E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 3.121E-05 | 7.977E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.764E-05 | 8.657E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.953E-05 | 8.265E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.411E-07 | 5.544E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.43E-06 | 1.644E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2E-06 | 1.533E-03 |
|---|
| interphase | GO:0051325 |  | 2.193E-06 | 1.261E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.307E-05 | 6.011E-03 |
|---|
| response to UV | GO:0009411 |  | 1.604E-05 | 6.149E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 7.697E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.121E-05 | 8.974E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 3.121E-05 | 7.977E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.764E-05 | 8.657E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.953E-05 | 8.265E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.411E-07 | 5.544E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.43E-06 | 1.644E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2E-06 | 1.533E-03 |
|---|
| interphase | GO:0051325 |  | 2.193E-06 | 1.261E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.307E-05 | 6.011E-03 |
|---|
| response to UV | GO:0009411 |  | 1.604E-05 | 6.149E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 7.697E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.121E-05 | 8.974E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 3.121E-05 | 7.977E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.764E-05 | 8.657E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.953E-05 | 8.265E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.411E-07 | 5.544E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.43E-06 | 1.644E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2E-06 | 1.533E-03 |
|---|
| interphase | GO:0051325 |  | 2.193E-06 | 1.261E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.307E-05 | 6.011E-03 |
|---|
| response to UV | GO:0009411 |  | 1.604E-05 | 6.149E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 7.697E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.121E-05 | 8.974E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 3.121E-05 | 7.977E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.764E-05 | 8.657E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.953E-05 | 8.265E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| detection of mechanical stimulus | GO:0050982 |  | 6.526E-10 | 1.549E-06 |
|---|
| cartilage condensation | GO:0001502 |  | 9.131E-10 | 1.083E-06 |
|---|
| cell cycle arrest | GO:0007050 |  | 3.847E-08 | 3.043E-05 |
|---|
| response to mechanical stimulus | GO:0009612 |  | 4.173E-08 | 2.475E-05 |
|---|
| JAK-STAT cascade | GO:0007259 |  | 1.213E-07 | 5.757E-05 |
|---|
| detection of abiotic stimulus | GO:0009582 |  | 1.734E-07 | 6.858E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.601E-07 | 8.816E-05 |
|---|
| cartilage development | GO:0051216 |  | 3.255E-07 | 9.656E-05 |
|---|
| skeletal system morphogenesis | GO:0048705 |  | 4.026E-07 | 1.061E-04 |
|---|
| detection of external stimulus | GO:0009581 |  | 4.924E-07 | 1.168E-04 |
|---|
| cell-matrix adhesion | GO:0007160 |  | 8.524E-07 | 1.839E-04 |
|---|



