Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 54805-6-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1616 | 1.044e-02 | 1.248e-02 | 1.023e-01 |
|---|
| IPC-NIBC-129 | 0.2298 | 9.560e-02 | 1.258e-01 | 6.588e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2462 | 3.700e-03 | 6.296e-03 | 1.394e-01 |
|---|
| Loi_GPL570 | 0.2893 | 8.087e-02 | 8.036e-02 | 3.588e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1730 | 6.780e-04 | 2.629e-03 | 2.406e-02 |
|---|
| Parker_GPL1390 | 0.2106 | 1.666e-01 | 1.729e-01 | 6.793e-01 |
|---|
| Parker_GPL887 | 0.0556 | 8.183e-01 | 8.359e-01 | 8.712e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3622 | 2.022e-01 | 1.653e-01 | 7.481e-01 |
|---|
| Schmidt | 0.1770 | 1.250e-02 | 1.769e-02 | 1.551e-01 |
|---|
| Sotiriou | 0.3015 | 8.187e-03 | 3.226e-03 | 1.423e-01 |
|---|
| Wang | 0.1761 | 8.987e-02 | 6.071e-02 | 3.407e-01 |
|---|
| Zhang | 0.2014 | 1.108e-01 | 7.402e-02 | 2.629e-01 |
|---|
| Zhou | 0.3410 | 6.003e-02 | 6.336e-02 | 3.723e-01 |
|---|
Expression data for subnetwork 54805-6-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
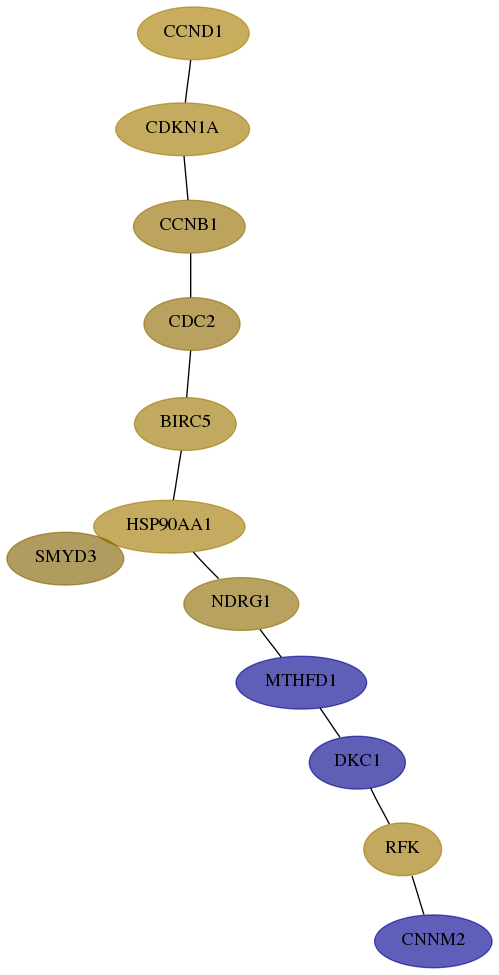
Score for each gene in subnetwork 54805-6-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| mthfd1 |   | 1 | 164 | 393 | 393 | -0.068 | 0.248 | 0.146 | 0.016 | 0.060 | 0.152 | 0.003 | 0.273 | 0.173 | -0.039 | 0.110 | 0.065 | 0.239 |
|---|
| dkc1 |   | 1 | 164 | 393 | 393 | -0.064 | 0.229 | 0.078 | 0.185 | 0.115 | 0.191 | -0.021 | 0.172 | 0.246 | 0.140 | 0.046 | 0.043 | 0.236 |
|---|
| ccnb1 |   | 36 | 7 | 78 | 59 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | 0.152 | 0.167 | 0.398 |
|---|
| hsp90aa1 |   | 11 | 21 | 234 | 204 | 0.116 | 0.030 | 0.164 | 0.076 | 0.119 | 0.155 | -0.199 | 0.169 | 0.069 | 0.102 | 0.052 | -0.011 | 0.399 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| ndrg1 |   | 9 | 27 | 250 | 223 | 0.072 | 0.301 | 0.084 | 0.324 | 0.108 | 0.102 | 0.240 | 0.083 | 0.068 | 0.064 | 0.128 | 0.092 | -0.255 |
|---|
| rfk |   | 1 | 164 | 393 | 393 | 0.106 | 0.039 | 0.004 | 0.303 | 0.122 | 0.008 | -0.068 | -0.100 | -0.058 | 0.056 | 0.079 | 0.070 | 0.120 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| cnnm2 |   | 1 | 164 | 393 | 393 | -0.077 | -0.037 | -0.103 | 0.179 | 0.073 | -0.176 | 0.485 | 0.159 | 0.081 | 0.027 | -0.032 | -0.002 | 0.129 |
|---|
| smyd3 |   | 1 | 164 | 393 | 393 | 0.054 | 0.070 | 0.040 | -0.209 | 0.087 | 0.021 | 0.200 | 0.096 | -0.119 | 0.016 | 0.089 | 0.105 | -0.043 |
|---|
| birc5 |   | 14 | 17 | 162 | 140 | 0.101 | 0.216 | 0.190 | 0.134 | 0.223 | 0.149 | 0.166 | 0.202 | 0.236 | 0.241 | 0.124 | 0.205 | 0.400 |
|---|
GO Enrichment output for subnetwork 54805-6-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.11E-08 | 2.552E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.315E-08 | 7.262E-05 |
|---|
| interphase | GO:0051325 |  | 7.092E-08 | 5.437E-05 |
|---|
| water-soluble vitamin biosynthetic process | GO:0042364 |  | 1.945E-06 | 1.118E-03 |
|---|
| vitamin biosynthetic process | GO:0009110 |  | 2.623E-06 | 1.206E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.066E-05 | 7.92E-03 |
|---|
| protein refolding | GO:0042026 |  | 2.261E-05 | 7.428E-03 |
|---|
| histidine metabolic process | GO:0006547 |  | 2.261E-05 | 6.5E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 2.261E-05 | 5.777E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 2.261E-05 | 5.2E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.372E-05 | 4.959E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.303E-09 | 8.068E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.106E-08 | 1.35E-05 |
|---|
| water-soluble vitamin biosynthetic process | GO:0042364 |  | 1.004E-06 | 8.18E-04 |
|---|
| vitamin biosynthetic process | GO:0009110 |  | 1.264E-06 | 7.722E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.805E-06 | 2.836E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 6.608E-06 | 2.691E-03 |
|---|
| protein refolding | GO:0042026 |  | 8.319E-06 | 2.903E-03 |
|---|
| response to UV | GO:0009411 |  | 9.988E-06 | 3.05E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.113E-05 | 3.021E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.164E-05 | 2.844E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.164E-05 | 2.586E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.615E-09 | 1.11E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.419E-08 | 1.707E-05 |
|---|
| interphase | GO:0051325 |  | 1.66E-08 | 1.332E-05 |
|---|
| water-soluble vitamin biosynthetic process | GO:0042364 |  | 1.297E-06 | 7.802E-04 |
|---|
| vitamin biosynthetic process | GO:0009110 |  | 1.633E-06 | 7.856E-04 |
|---|
| histidine catabolic process | GO:0006548 |  | 6.605E-06 | 2.649E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.491E-06 | 2.575E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.526E-06 | 2.564E-03 |
|---|
| protein refolding | GO:0042026 |  | 9.903E-06 | 2.647E-03 |
|---|
| response to UV | GO:0009411 |  | 1.152E-05 | 2.771E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.386E-05 | 3.031E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.303E-09 | 8.068E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.106E-08 | 1.35E-05 |
|---|
| water-soluble vitamin biosynthetic process | GO:0042364 |  | 1.004E-06 | 8.18E-04 |
|---|
| vitamin biosynthetic process | GO:0009110 |  | 1.264E-06 | 7.722E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.805E-06 | 2.836E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 6.608E-06 | 2.691E-03 |
|---|
| protein refolding | GO:0042026 |  | 8.319E-06 | 2.903E-03 |
|---|
| response to UV | GO:0009411 |  | 9.988E-06 | 3.05E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.113E-05 | 3.021E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.164E-05 | 2.844E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.164E-05 | 2.586E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.615E-09 | 1.11E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.419E-08 | 1.707E-05 |
|---|
| interphase | GO:0051325 |  | 1.66E-08 | 1.332E-05 |
|---|
| water-soluble vitamin biosynthetic process | GO:0042364 |  | 1.297E-06 | 7.802E-04 |
|---|
| vitamin biosynthetic process | GO:0009110 |  | 1.633E-06 | 7.856E-04 |
|---|
| histidine catabolic process | GO:0006548 |  | 6.605E-06 | 2.649E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.491E-06 | 2.575E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.526E-06 | 2.564E-03 |
|---|
| protein refolding | GO:0042026 |  | 9.903E-06 | 2.647E-03 |
|---|
| response to UV | GO:0009411 |  | 1.152E-05 | 2.771E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.386E-05 | 3.031E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.259E-08 | 4.494E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 3.704E-08 | 6.614E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.523E-07 | 1.813E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.824E-07 | 1.629E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.366E-06 | 2.404E-03 |
|---|
| response to UV | GO:0009411 |  | 4.081E-06 | 2.429E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 4.142E-06 | 2.113E-03 |
|---|
| formate-tetrahydrofolate ligase activity | GO:0004329 |  | 4.142E-06 | 1.849E-03 |
|---|
| nitric-oxide synthase regulator activity | GO:0030235 |  | 4.142E-06 | 1.643E-03 |
|---|
| cyclohydrolase activity | GO:0019238 |  | 4.142E-06 | 1.479E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.339E-06 | 1.409E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.754E-10 | 6.398E-07 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.035E-09 | 1.888E-06 |
|---|
| interphase | GO:0051325 |  | 1.172E-09 | 1.425E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.125E-08 | 1.026E-05 |
|---|
| midbody | GO:0030496 |  | 4.413E-08 | 3.22E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 7.296E-08 | 4.436E-05 |
|---|
| spindle microtubule | GO:0005876 |  | 1.633E-07 | 8.512E-05 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.448E-06 | 1.116E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.448E-06 | 9.922E-04 |
|---|
| response to UV | GO:0009411 |  | 2.813E-06 | 1.026E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 3.763E-06 | 1.248E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.615E-09 | 1.11E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.419E-08 | 1.707E-05 |
|---|
| interphase | GO:0051325 |  | 1.66E-08 | 1.332E-05 |
|---|
| water-soluble vitamin biosynthetic process | GO:0042364 |  | 1.297E-06 | 7.802E-04 |
|---|
| vitamin biosynthetic process | GO:0009110 |  | 1.633E-06 | 7.856E-04 |
|---|
| histidine catabolic process | GO:0006548 |  | 6.605E-06 | 2.649E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.491E-06 | 2.575E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.526E-06 | 2.564E-03 |
|---|
| protein refolding | GO:0042026 |  | 9.903E-06 | 2.647E-03 |
|---|
| response to UV | GO:0009411 |  | 1.152E-05 | 2.771E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.386E-05 | 3.031E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.11E-08 | 2.552E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.315E-08 | 7.262E-05 |
|---|
| interphase | GO:0051325 |  | 7.092E-08 | 5.437E-05 |
|---|
| water-soluble vitamin biosynthetic process | GO:0042364 |  | 1.945E-06 | 1.118E-03 |
|---|
| vitamin biosynthetic process | GO:0009110 |  | 2.623E-06 | 1.206E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.066E-05 | 7.92E-03 |
|---|
| protein refolding | GO:0042026 |  | 2.261E-05 | 7.428E-03 |
|---|
| histidine metabolic process | GO:0006547 |  | 2.261E-05 | 6.5E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 2.261E-05 | 5.777E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 2.261E-05 | 5.2E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.372E-05 | 4.959E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.11E-08 | 2.552E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.315E-08 | 7.262E-05 |
|---|
| interphase | GO:0051325 |  | 7.092E-08 | 5.437E-05 |
|---|
| water-soluble vitamin biosynthetic process | GO:0042364 |  | 1.945E-06 | 1.118E-03 |
|---|
| vitamin biosynthetic process | GO:0009110 |  | 2.623E-06 | 1.206E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.066E-05 | 7.92E-03 |
|---|
| protein refolding | GO:0042026 |  | 2.261E-05 | 7.428E-03 |
|---|
| histidine metabolic process | GO:0006547 |  | 2.261E-05 | 6.5E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 2.261E-05 | 5.777E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 2.261E-05 | 5.2E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.372E-05 | 4.959E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.11E-08 | 2.552E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.315E-08 | 7.262E-05 |
|---|
| interphase | GO:0051325 |  | 7.092E-08 | 5.437E-05 |
|---|
| water-soluble vitamin biosynthetic process | GO:0042364 |  | 1.945E-06 | 1.118E-03 |
|---|
| vitamin biosynthetic process | GO:0009110 |  | 2.623E-06 | 1.206E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.066E-05 | 7.92E-03 |
|---|
| protein refolding | GO:0042026 |  | 2.261E-05 | 7.428E-03 |
|---|
| histidine metabolic process | GO:0006547 |  | 2.261E-05 | 6.5E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 2.261E-05 | 5.777E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 2.261E-05 | 5.2E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.372E-05 | 4.959E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.11E-08 | 2.552E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.315E-08 | 7.262E-05 |
|---|
| interphase | GO:0051325 |  | 7.092E-08 | 5.437E-05 |
|---|
| water-soluble vitamin biosynthetic process | GO:0042364 |  | 1.945E-06 | 1.118E-03 |
|---|
| vitamin biosynthetic process | GO:0009110 |  | 2.623E-06 | 1.206E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.066E-05 | 7.92E-03 |
|---|
| protein refolding | GO:0042026 |  | 2.261E-05 | 7.428E-03 |
|---|
| histidine metabolic process | GO:0006547 |  | 2.261E-05 | 6.5E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 2.261E-05 | 5.777E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 2.261E-05 | 5.2E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.372E-05 | 4.959E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 9.626E-09 | 2.284E-05 |
|---|
| protein refolding | GO:0042026 |  | 3.081E-08 | 3.655E-05 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 5.387E-08 | 4.261E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.677E-08 | 3.368E-05 |
|---|
| interphase | GO:0051325 |  | 6.367E-08 | 3.022E-05 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 8.612E-08 | 3.406E-05 |
|---|
| pseudouridine synthesis | GO:0001522 |  | 1.291E-07 | 4.376E-05 |
|---|
| telomere maintenance via telomerase | GO:0007004 |  | 1.843E-07 | 5.466E-05 |
|---|
| RNA-dependent DNA replication | GO:0006278 |  | 3.373E-07 | 8.893E-05 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.737E-06 | 4.121E-04 |
|---|
| water-soluble vitamin biosynthetic process | GO:0042364 |  | 2.342E-06 | 5.053E-04 |
|---|



