Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 5315-6-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1714 | 9.742e-03 | 1.164e-02 | 9.716e-02 |
|---|
| IPC-NIBC-129 | 0.2526 | 6.723e-02 | 8.705e-02 | 5.444e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2496 | 8.673e-03 | 1.354e-02 | 2.416e-01 |
|---|
| Loi_GPL570 | 0.2496 | 5.934e-02 | 5.892e-02 | 2.887e-01 |
|---|
| Loi_GPL96-GPL97 | 0.2099 | 3.570e-04 | 1.507e-03 | 1.118e-02 |
|---|
| Parker_GPL1390 | 0.2366 | 1.458e-01 | 1.498e-01 | 6.423e-01 |
|---|
| Parker_GPL887 | 0.0675 | 7.716e-01 | 7.926e-01 | 8.272e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3441 | 2.500e-01 | 2.111e-01 | 8.138e-01 |
|---|
| Schmidt | 0.1868 | 1.119e-02 | 1.591e-02 | 1.426e-01 |
|---|
| Sotiriou | 0.3083 | 4.593e-03 | 1.558e-03 | 9.596e-02 |
|---|
| Wang | 0.1570 | 4.922e-02 | 3.005e-02 | 2.253e-01 |
|---|
| Zhang | 0.1796 | 1.454e-01 | 1.025e-01 | 3.320e-01 |
|---|
| Zhou | 0.3762 | 7.133e-02 | 7.651e-02 | 4.292e-01 |
|---|
Expression data for subnetwork 5315-6-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
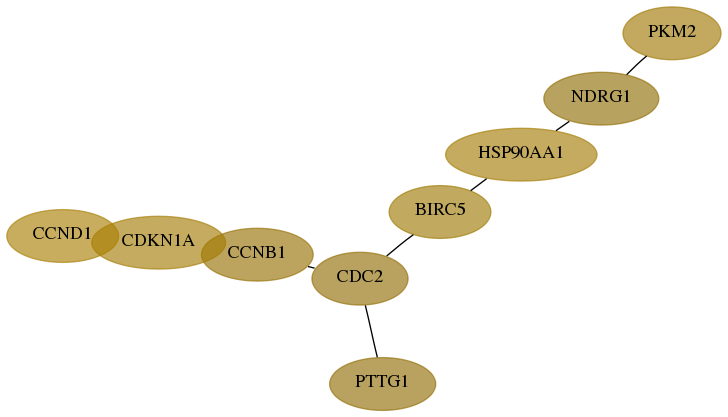
Score for each gene in subnetwork 5315-6-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| pkm2 |   | 1 | 164 | 390 | 390 | 0.103 | 0.094 | 0.089 | -0.216 | 0.079 | 0.197 | -0.500 | 0.029 | -0.027 | 0.057 | -0.033 | -0.099 | 0.166 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| pttg1 |   | 10 | 25 | 31 | 21 | 0.072 | 0.270 | 0.175 | 0.183 | 0.200 | 0.208 | 0.347 | 0.334 | 0.224 | 0.204 | 0.147 | 0.210 | 0.309 |
|---|
| ccnb1 |   | 36 | 7 | 78 | 59 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | 0.152 | 0.167 | 0.398 |
|---|
| hsp90aa1 |   | 11 | 21 | 234 | 204 | 0.116 | 0.030 | 0.164 | 0.076 | 0.119 | 0.155 | -0.199 | 0.169 | 0.069 | 0.102 | 0.052 | -0.011 | 0.399 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| ndrg1 |   | 9 | 27 | 250 | 223 | 0.072 | 0.301 | 0.084 | 0.324 | 0.108 | 0.102 | 0.240 | 0.083 | 0.068 | 0.064 | 0.128 | 0.092 | -0.255 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| birc5 |   | 14 | 17 | 162 | 140 | 0.101 | 0.216 | 0.190 | 0.134 | 0.223 | 0.149 | 0.166 | 0.202 | 0.236 | 0.241 | 0.124 | 0.205 | 0.400 |
|---|
GO Enrichment output for subnetwork 5315-6-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.656E-09 | 1.071E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.087E-08 | 2.399E-05 |
|---|
| interphase | GO:0051325 |  | 2.344E-08 | 1.797E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.102E-05 | 6.338E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.266E-05 | 5.823E-03 |
|---|
| response to UV | GO:0009411 |  | 1.353E-05 | 5.188E-03 |
|---|
| protein refolding | GO:0042026 |  | 1.498E-05 | 4.923E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.498E-05 | 4.308E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 1.498E-05 | 3.829E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.851E-05 | 4.256E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.096E-05 | 4.383E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 9.148E-10 | 2.235E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.156E-09 | 2.633E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.277E-06 | 1.854E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.593E-06 | 1.583E-03 |
|---|
| response to UV | GO:0009411 |  | 3.923E-06 | 1.917E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.372E-06 | 1.78E-03 |
|---|
| protein refolding | GO:0042026 |  | 4.506E-06 | 1.573E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 6.306E-06 | 1.926E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 6.306E-06 | 1.712E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 7.458E-06 | 1.822E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 7.795E-06 | 1.731E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.772E-09 | 4.264E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.192E-09 | 5.043E-06 |
|---|
| interphase | GO:0051325 |  | 4.908E-09 | 3.936E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.733E-06 | 2.245E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.249E-06 | 2.045E-03 |
|---|
| response to UV | GO:0009411 |  | 5.743E-06 | 2.303E-03 |
|---|
| protein refolding | GO:0042026 |  | 6.274E-06 | 2.157E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.162E-06 | 2.154E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 8.781E-06 | 2.347E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 8.781E-06 | 2.113E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.17E-05 | 2.56E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 9.148E-10 | 2.235E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.156E-09 | 2.633E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.277E-06 | 1.854E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 2.593E-06 | 1.583E-03 |
|---|
| response to UV | GO:0009411 |  | 3.923E-06 | 1.917E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.372E-06 | 1.78E-03 |
|---|
| protein refolding | GO:0042026 |  | 4.506E-06 | 1.573E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 6.306E-06 | 1.926E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 6.306E-06 | 1.712E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 7.458E-06 | 1.822E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 7.795E-06 | 1.731E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.772E-09 | 4.264E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.192E-09 | 5.043E-06 |
|---|
| interphase | GO:0051325 |  | 4.908E-09 | 3.936E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.733E-06 | 2.245E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.249E-06 | 2.045E-03 |
|---|
| response to UV | GO:0009411 |  | 5.743E-06 | 2.303E-03 |
|---|
| protein refolding | GO:0042026 |  | 6.274E-06 | 2.157E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.162E-06 | 2.154E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 8.781E-06 | 2.347E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 8.781E-06 | 2.113E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.17E-05 | 2.56E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 5.876E-09 | 2.098E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.73E-08 | 3.089E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.132E-08 | 7.3E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 7.119E-08 | 6.356E-05 |
|---|
| cysteine-type endopeptidase inhibitor activity | GO:0004869 |  | 4.236E-07 | 3.026E-04 |
|---|
| regulation of Rab protein signal transduction | GO:0032483 |  | 1.028E-06 | 6.12E-04 |
|---|
| Rab GTPase activator activity | GO:0005097 |  | 1.109E-06 | 5.659E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.578E-06 | 7.042E-04 |
|---|
| response to UV | GO:0009411 |  | 1.913E-06 | 7.591E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.035E-06 | 7.266E-04 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 2.578E-06 | 8.368E-04 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 7.449E-11 | 2.717E-07 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.315E-10 | 6.047E-07 |
|---|
| interphase | GO:0051325 |  | 3.755E-10 | 4.566E-07 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 6.139E-09 | 5.598E-06 |
|---|
| midbody | GO:0030496 |  | 2.409E-08 | 1.758E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 3.983E-08 | 2.422E-05 |
|---|
| spindle microtubule | GO:0005876 |  | 8.92E-08 | 4.648E-05 |
|---|
| cysteine-type endopeptidase inhibitor activity | GO:0004869 |  | 5.939E-07 | 2.708E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.34E-06 | 5.43E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.34E-06 | 4.887E-04 |
|---|
| response to UV | GO:0009411 |  | 1.539E-06 | 5.105E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.772E-09 | 4.264E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.192E-09 | 5.043E-06 |
|---|
| interphase | GO:0051325 |  | 4.908E-09 | 3.936E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.733E-06 | 2.245E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.249E-06 | 2.045E-03 |
|---|
| response to UV | GO:0009411 |  | 5.743E-06 | 2.303E-03 |
|---|
| protein refolding | GO:0042026 |  | 6.274E-06 | 2.157E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.162E-06 | 2.154E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 8.781E-06 | 2.347E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 8.781E-06 | 2.113E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.17E-05 | 2.56E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.656E-09 | 1.071E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.087E-08 | 2.399E-05 |
|---|
| interphase | GO:0051325 |  | 2.344E-08 | 1.797E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.102E-05 | 6.338E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.266E-05 | 5.823E-03 |
|---|
| response to UV | GO:0009411 |  | 1.353E-05 | 5.188E-03 |
|---|
| protein refolding | GO:0042026 |  | 1.498E-05 | 4.923E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.498E-05 | 4.308E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 1.498E-05 | 3.829E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.851E-05 | 4.256E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.096E-05 | 4.383E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.656E-09 | 1.071E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.087E-08 | 2.399E-05 |
|---|
| interphase | GO:0051325 |  | 2.344E-08 | 1.797E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.102E-05 | 6.338E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.266E-05 | 5.823E-03 |
|---|
| response to UV | GO:0009411 |  | 1.353E-05 | 5.188E-03 |
|---|
| protein refolding | GO:0042026 |  | 1.498E-05 | 4.923E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.498E-05 | 4.308E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 1.498E-05 | 3.829E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.851E-05 | 4.256E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.096E-05 | 4.383E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.656E-09 | 1.071E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.087E-08 | 2.399E-05 |
|---|
| interphase | GO:0051325 |  | 2.344E-08 | 1.797E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.102E-05 | 6.338E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.266E-05 | 5.823E-03 |
|---|
| response to UV | GO:0009411 |  | 1.353E-05 | 5.188E-03 |
|---|
| protein refolding | GO:0042026 |  | 1.498E-05 | 4.923E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.498E-05 | 4.308E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 1.498E-05 | 3.829E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.851E-05 | 4.256E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.096E-05 | 4.383E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.656E-09 | 1.071E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.087E-08 | 2.399E-05 |
|---|
| interphase | GO:0051325 |  | 2.344E-08 | 1.797E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.102E-05 | 6.338E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.266E-05 | 5.823E-03 |
|---|
| response to UV | GO:0009411 |  | 1.353E-05 | 5.188E-03 |
|---|
| protein refolding | GO:0042026 |  | 1.498E-05 | 4.923E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.498E-05 | 4.308E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 1.498E-05 | 3.829E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.851E-05 | 4.256E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.096E-05 | 4.383E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.523E-09 | 8.361E-06 |
|---|
| protein refolding | GO:0042026 |  | 1.476E-08 | 1.751E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.58E-08 | 1.25E-05 |
|---|
| interphase | GO:0051325 |  | 1.773E-08 | 1.052E-05 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 2.581E-08 | 1.225E-05 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 4.127E-08 | 1.632E-05 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 8.341E-07 | 2.828E-04 |
|---|
| response to unfolded protein | GO:0006986 |  | 8.985E-07 | 2.665E-04 |
|---|
| response to protein stimulus | GO:0051789 |  | 1.147E-06 | 3.024E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.477E-06 | 3.506E-04 |
|---|
| mitochondrial membrane organization | GO:0007006 |  | 1.678E-06 | 3.619E-04 |
|---|



