Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 4331-5-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1569 | 9.310e-03 | 1.112e-02 | 9.391e-02 |
|---|
| IPC-NIBC-129 | 0.2523 | 5.240e-02 | 6.691e-02 | 4.665e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2518 | 1.640e-02 | 2.403e-02 | 3.498e-01 |
|---|
| Loi_GPL570 | 0.2197 | 5.960e-02 | 5.918e-02 | 2.897e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1710 | 6.000e-05 | 3.180e-04 | 1.069e-03 |
|---|
| Parker_GPL1390 | 0.2549 | 1.774e-01 | 1.850e-01 | 6.968e-01 |
|---|
| Parker_GPL887 | 0.1212 | 5.757e-01 | 6.070e-01 | 6.209e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3547 | 1.318e-01 | 1.007e-01 | 6.074e-01 |
|---|
| Schmidt | 0.2131 | 1.036e-02 | 1.478e-02 | 1.344e-01 |
|---|
| Sotiriou | 0.3130 | 3.012e-03 | 9.150e-04 | 7.125e-02 |
|---|
| Wang | 0.1940 | 9.270e-02 | 6.295e-02 | 3.478e-01 |
|---|
| Zhang | 0.2441 | 8.562e-02 | 5.439e-02 | 2.092e-01 |
|---|
| Zhou | 0.4017 | 6.449e-02 | 6.854e-02 | 3.954e-01 |
|---|
Expression data for subnetwork 4331-5-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
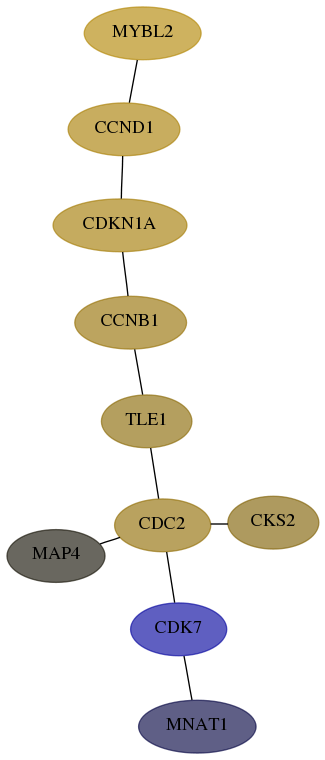
Score for each gene in subnetwork 4331-5-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| mybl2 |   | 3 | 79 | 44 | 44 | 0.162 | 0.237 | 0.166 | 0.079 | 0.124 | 0.184 | 0.333 | 0.275 | 0.202 | 0.182 | 0.120 | 0.074 | 0.245 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| tle1 |   | 23 | 9 | 78 | 60 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | -0.028 | 0.068 | 0.025 |
|---|
| ccnb1 |   | 36 | 7 | 78 | 59 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | 0.152 | 0.167 | 0.398 |
|---|
| cdk7 |   | 2 | 104 | 185 | 179 | -0.102 | 0.121 | 0.116 | 0.126 | 0.170 | 0.008 | -0.135 | 0.090 | -0.085 | 0.182 | -0.023 | 0.021 | 0.168 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| cks2 |   | 44 | 6 | 73 | 54 | 0.050 | 0.233 | 0.170 | 0.103 | 0.186 | 0.172 | 0.093 | 0.167 | 0.263 | 0.151 | 0.145 | 0.284 | 0.383 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| map4 |   | 16 | 13 | 73 | 55 | 0.002 | 0.046 | 0.067 | -0.043 | 0.034 | 0.119 | 0.159 | -0.058 | -0.020 | 0.043 | 0.108 | -0.074 | 0.302 |
|---|
| mnat1 |   | 3 | 79 | 175 | 162 | -0.011 | 0.083 | 0.075 | 0.305 | 0.105 | 0.111 | -0.009 | 0.107 | 0.118 | -0.002 | 0.144 | 0.307 | 0.086 |
|---|
GO Enrichment output for subnetwork 4331-5-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.387E-11 | 5.489E-08 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.959E-06 | 2.253E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.959E-06 | 1.502E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.065E-06 | 1.762E-03 |
|---|
| interphase | GO:0051325 |  | 3.36E-06 | 1.546E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 1.434E-05 | 5.498E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.787E-05 | 5.873E-03 |
|---|
| response to UV | GO:0009411 |  | 2.194E-05 | 6.306E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 2.341E-05 | 5.983E-03 |
|---|
| RNA elongation | GO:0006354 |  | 2.656E-05 | 6.109E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 6.013E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.753E-12 | 1.161E-08 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 7.671E-07 | 9.37E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 9.952E-07 | 8.104E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.057E-06 | 6.457E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.496E-06 | 3.174E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 7.872E-06 | 3.205E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 9.428E-06 | 3.29E-03 |
|---|
| response to UV | GO:0009411 |  | 1.117E-05 | 3.412E-03 |
|---|
| RNA elongation | GO:0006354 |  | 1.18E-05 | 3.203E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.245E-05 | 3.041E-03 |
|---|
| DNA catabolic process | GO:0006308 |  | 1.454E-05 | 3.228E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.006E-11 | 2.42E-08 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.124E-06 | 1.353E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.458E-06 | 1.17E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.542E-06 | 9.275E-04 |
|---|
| interphase | GO:0051325 |  | 1.746E-06 | 8.404E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.506E-06 | 3.812E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 1.015E-05 | 3.488E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 1.3E-05 | 3.91E-03 |
|---|
| response to UV | GO:0009411 |  | 1.461E-05 | 3.905E-03 |
|---|
| RNA elongation | GO:0006354 |  | 1.634E-05 | 3.932E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.726E-05 | 3.774E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.753E-12 | 1.161E-08 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 7.671E-07 | 9.37E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 9.952E-07 | 8.104E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.057E-06 | 6.457E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.496E-06 | 3.174E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 7.872E-06 | 3.205E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 9.428E-06 | 3.29E-03 |
|---|
| response to UV | GO:0009411 |  | 1.117E-05 | 3.412E-03 |
|---|
| RNA elongation | GO:0006354 |  | 1.18E-05 | 3.203E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.245E-05 | 3.041E-03 |
|---|
| DNA catabolic process | GO:0006308 |  | 1.454E-05 | 3.228E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.006E-11 | 2.42E-08 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.124E-06 | 1.353E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.458E-06 | 1.17E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.542E-06 | 9.275E-04 |
|---|
| interphase | GO:0051325 |  | 1.746E-06 | 8.404E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.506E-06 | 3.812E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 1.015E-05 | 3.488E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 1.3E-05 | 3.91E-03 |
|---|
| response to UV | GO:0009411 |  | 1.461E-05 | 3.905E-03 |
|---|
| RNA elongation | GO:0006354 |  | 1.634E-05 | 3.932E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.726E-05 | 3.774E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.327E-13 | 8.309E-10 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 3.522E-11 | 6.289E-08 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.259E-08 | 1.498E-05 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 6.666E-08 | 5.951E-05 |
|---|
| kinase regulator activity | GO:0019207 |  | 1.414E-07 | 1.01E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.523E-07 | 9.066E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.824E-07 | 9.306E-05 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 3.959E-07 | 1.767E-04 |
|---|
| response to calcium ion | GO:0051592 |  | 1.457E-06 | 5.78E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.366E-06 | 1.202E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 3.366E-06 | 1.093E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.379E-13 | 5.03E-10 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 6.438E-11 | 1.174E-07 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 1.508E-09 | 1.834E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 8.439E-09 | 7.696E-06 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 4.564E-08 | 3.33E-05 |
|---|
| kinase regulator activity | GO:0019207 |  | 8.1E-08 | 4.925E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 8.417E-08 | 4.387E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.049E-07 | 4.783E-05 |
|---|
| interphase | GO:0051325 |  | 1.158E-07 | 4.693E-05 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.996E-07 | 7.281E-05 |
|---|
| cyclin-dependent protein kinase activity | GO:0004693 |  | 4.381E-07 | 1.453E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.006E-11 | 2.42E-08 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.124E-06 | 1.353E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.458E-06 | 1.17E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.542E-06 | 9.275E-04 |
|---|
| interphase | GO:0051325 |  | 1.746E-06 | 8.404E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.506E-06 | 3.812E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 1.015E-05 | 3.488E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 1.3E-05 | 3.91E-03 |
|---|
| response to UV | GO:0009411 |  | 1.461E-05 | 3.905E-03 |
|---|
| RNA elongation | GO:0006354 |  | 1.634E-05 | 3.932E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 1.726E-05 | 3.774E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.387E-11 | 5.489E-08 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.959E-06 | 2.253E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.959E-06 | 1.502E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.065E-06 | 1.762E-03 |
|---|
| interphase | GO:0051325 |  | 3.36E-06 | 1.546E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 1.434E-05 | 5.498E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.787E-05 | 5.873E-03 |
|---|
| response to UV | GO:0009411 |  | 2.194E-05 | 6.306E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 2.341E-05 | 5.983E-03 |
|---|
| RNA elongation | GO:0006354 |  | 2.656E-05 | 6.109E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 6.013E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.387E-11 | 5.489E-08 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.959E-06 | 2.253E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.959E-06 | 1.502E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.065E-06 | 1.762E-03 |
|---|
| interphase | GO:0051325 |  | 3.36E-06 | 1.546E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 1.434E-05 | 5.498E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.787E-05 | 5.873E-03 |
|---|
| response to UV | GO:0009411 |  | 2.194E-05 | 6.306E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 2.341E-05 | 5.983E-03 |
|---|
| RNA elongation | GO:0006354 |  | 2.656E-05 | 6.109E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 6.013E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.387E-11 | 5.489E-08 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.959E-06 | 2.253E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.959E-06 | 1.502E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.065E-06 | 1.762E-03 |
|---|
| interphase | GO:0051325 |  | 3.36E-06 | 1.546E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 1.434E-05 | 5.498E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.787E-05 | 5.873E-03 |
|---|
| response to UV | GO:0009411 |  | 2.194E-05 | 6.306E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 2.341E-05 | 5.983E-03 |
|---|
| RNA elongation | GO:0006354 |  | 2.656E-05 | 6.109E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 6.013E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.387E-11 | 5.489E-08 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.959E-06 | 2.253E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.959E-06 | 1.502E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.065E-06 | 1.762E-03 |
|---|
| interphase | GO:0051325 |  | 3.36E-06 | 1.546E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 1.434E-05 | 5.498E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.787E-05 | 5.873E-03 |
|---|
| response to UV | GO:0009411 |  | 2.194E-05 | 6.306E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 2.341E-05 | 5.983E-03 |
|---|
| RNA elongation | GO:0006354 |  | 2.656E-05 | 6.109E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 6.013E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.407E-11 | 3.339E-08 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.333E-06 | 1.582E-03 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.542E-06 | 1.22E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.931E-06 | 1.146E-03 |
|---|
| interphase | GO:0051325 |  | 2.116E-06 | 1.004E-03 |
|---|
| response to calcium ion | GO:0051592 |  | 1.219E-05 | 4.819E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.307E-05 | 4.43E-03 |
|---|
| response to UV | GO:0009411 |  | 1.597E-05 | 4.736E-03 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 1.812E-05 | 4.777E-03 |
|---|
| RNA elongation | GO:0006354 |  | 2.045E-05 | 4.853E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.223E-05 | 4.795E-03 |
|---|



