Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 2997-4-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2244 | 5.955e-03 | 7.089e-03 | 6.679e-02 |
|---|
| IPC-NIBC-129 | 0.2441 | 6.326e-02 | 8.165e-02 | 5.250e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2734 | 1.254e-02 | 1.887e-02 | 3.007e-01 |
|---|
| Loi_GPL570 | 0.2324 | 6.665e-02 | 6.620e-02 | 3.137e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1725 | 7.870e-04 | 2.995e-03 | 2.859e-02 |
|---|
| Parker_GPL1390 | 0.2411 | 6.984e-02 | 6.739e-02 | 4.515e-01 |
|---|
| Parker_GPL887 | 0.0656 | 7.791e-01 | 7.995e-01 | 8.344e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3568 | 1.804e-01 | 1.449e-01 | 7.112e-01 |
|---|
| Schmidt | 0.1747 | 1.360e-02 | 1.917e-02 | 1.652e-01 |
|---|
| Sotiriou | 0.2964 | 1.725e-03 | 4.530e-04 | 4.754e-02 |
|---|
| Wang | 0.2013 | 9.050e-02 | 6.120e-02 | 3.423e-01 |
|---|
| Zhang | 0.2128 | 7.704e-02 | 4.793e-02 | 1.901e-01 |
|---|
| Zhou | 0.4351 | 6.323e-02 | 6.707e-02 | 3.890e-01 |
|---|
Expression data for subnetwork 2997-4-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
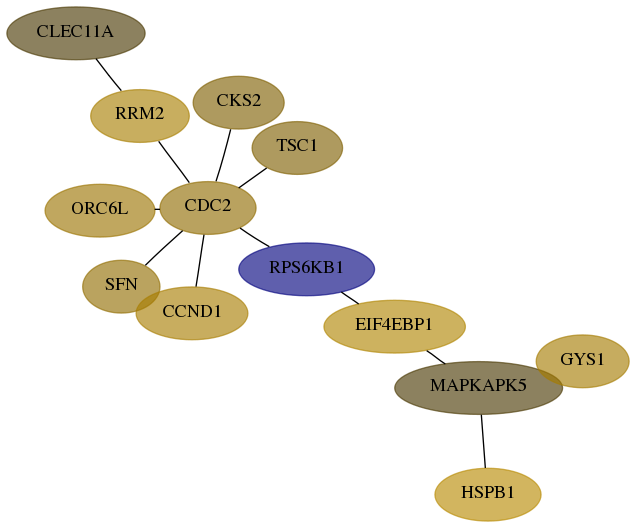
Score for each gene in subnetwork 2997-4-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| rps6kb1 |   | 2 | 104 | 307 | 295 | -0.048 | 0.004 | 0.033 | 0.144 | 0.128 | 0.095 | -0.319 | 0.022 | 0.116 | 0.084 | 0.184 | 0.076 | 0.280 |
|---|
| rrm2 |   | 21 | 10 | 44 | 26 | 0.131 | 0.207 | 0.227 | 0.187 | 0.161 | 0.178 | 0.137 | 0.303 | 0.190 | 0.219 | 0.117 | 0.129 | 0.302 |
|---|
| eif4ebp1 |   | 2 | 104 | 307 | 295 | 0.158 | 0.203 | 0.132 | 0.139 | 0.104 | 0.122 | 0.119 | 0.252 | 0.031 | 0.042 | 0.104 | 0.029 | 0.219 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| cks2 |   | 44 | 6 | 73 | 54 | 0.050 | 0.233 | 0.170 | 0.103 | 0.186 | 0.172 | 0.093 | 0.167 | 0.263 | 0.151 | 0.145 | 0.284 | 0.383 |
|---|
| hspb1 |   | 19 | 11 | 17 | 11 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | -0.026 | -0.065 | 0.289 | -0.021 | 0.201 | 0.125 | -0.077 | 0.290 |
|---|
| tsc1 |   | 47 | 5 | 17 | 8 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.043 | -0.007 | 0.207 |
|---|
| mapkapk5 |   | 1 | 164 | 347 | 347 | 0.013 | 0.083 | 0.079 | -0.082 | 0.107 | 0.011 | 0.630 | 0.269 | -0.066 | 0.073 | -0.002 | -0.034 | 0.191 |
|---|
| orc6l |   | 8 | 33 | 73 | 61 | 0.094 | 0.270 | 0.084 | 0.114 | 0.106 | 0.240 | 0.256 | 0.234 | 0.192 | 0.105 | 0.086 | 0.175 | 0.160 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| sfn |   | 50 | 4 | 29 | 17 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.025 | 0.079 | 0.181 |
|---|
| clec11a |   | 17 | 12 | 44 | 27 | 0.013 | 0.101 | 0.071 | -0.085 | 0.066 | 0.082 | 0.209 | -0.126 | -0.034 | -0.001 | 0.191 | 0.059 | -0.075 |
|---|
| gys1 |   | 1 | 164 | 347 | 347 | 0.119 | 0.142 | 0.070 | -0.156 | 0.067 | -0.053 | -0.221 | 0.212 | -0.070 | 0.098 | -0.004 | 0.025 | 0.289 |
|---|
GO Enrichment output for subnetwork 2997-4-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.981E-08 | 6.856E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 5.212E-08 | 5.994E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.706E-07 | 2.841E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 8.282E-07 | 4.762E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.959E-06 | 9.013E-04 |
|---|
| regulation of translation | GO:0006417 |  | 5.598E-06 | 2.146E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.371E-05 | 4.506E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.371E-05 | 3.943E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 2.052E-05 | 5.244E-03 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 2.056E-05 | 4.728E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 6.013E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.253E-08 | 1.039E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 2.33E-07 | 2.846E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.653E-06 | 2.975E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 5.001E-06 | 3.054E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.477E-06 | 2.676E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 7.665E-06 | 3.121E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.022E-05 | 3.566E-03 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 1.022E-05 | 3.12E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.022E-05 | 2.773E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 1.313E-05 | 3.208E-03 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.313E-05 | 2.916E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.359E-08 | 3.269E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 3.258E-08 | 3.919E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.936E-08 | 7.969E-05 |
|---|
| negative regulation of translation | GO:0017148 |  | 4.397E-07 | 2.645E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 5.935E-07 | 2.856E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.571E-06 | 2.234E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.571E-06 | 1.915E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 8.889E-06 | 2.673E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.558E-05 | 4.164E-03 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 1.558E-05 | 3.748E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.558E-05 | 3.407E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.253E-08 | 1.039E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 2.33E-07 | 2.846E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 3.653E-06 | 2.975E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 5.001E-06 | 3.054E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.477E-06 | 2.676E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 7.665E-06 | 3.121E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.022E-05 | 3.566E-03 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 1.022E-05 | 3.12E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.022E-05 | 2.773E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 1.313E-05 | 3.208E-03 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.313E-05 | 2.916E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.359E-08 | 3.269E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 3.258E-08 | 3.919E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.936E-08 | 7.969E-05 |
|---|
| negative regulation of translation | GO:0017148 |  | 4.397E-07 | 2.645E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 5.935E-07 | 2.856E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.571E-06 | 2.234E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.571E-06 | 1.915E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 8.889E-06 | 2.673E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.558E-05 | 4.164E-03 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 1.558E-05 | 3.748E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.558E-05 | 3.407E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.693E-08 | 2.033E-04 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 6.786E-08 | 1.212E-04 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 1.561E-07 | 1.858E-04 |
|---|
| kinase regulator activity | GO:0019207 |  | 3.306E-07 | 2.951E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 3.345E-07 | 2.389E-04 |
|---|
| regulation of translational initiation | GO:0006446 |  | 4.324E-06 | 2.574E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.073E-06 | 3.098E-03 |
|---|
| UDP-glucosyltransferase activity | GO:0035251 |  | 6.073E-06 | 2.711E-03 |
|---|
| eukaryotic initiation factor 4E binding | GO:0008190 |  | 6.073E-06 | 2.409E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.105E-06 | 3.251E-03 |
|---|
| nuclear origin of replication recognition complex | GO:0005664 |  | 9.105E-06 | 2.956E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.43E-08 | 1.251E-04 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 9.481E-08 | 1.729E-04 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 9.836E-08 | 1.196E-04 |
|---|
| kinase regulator activity | GO:0019207 |  | 1.744E-07 | 1.591E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 2.122E-07 | 1.548E-04 |
|---|
| regulation of translational initiation | GO:0006446 |  | 2.176E-06 | 1.323E-03 |
|---|
| response to insulin stimulus | GO:0032868 |  | 4.168E-06 | 2.172E-03 |
|---|
| origin recognition complex | GO:0000808 |  | 4.447E-06 | 2.028E-03 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 4.447E-06 | 1.802E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 4.447E-06 | 1.622E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.667E-06 | 2.211E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.359E-08 | 3.269E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 3.258E-08 | 3.919E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.936E-08 | 7.969E-05 |
|---|
| negative regulation of translation | GO:0017148 |  | 4.397E-07 | 2.645E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 5.935E-07 | 2.856E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.571E-06 | 2.234E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.571E-06 | 1.915E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 8.889E-06 | 2.673E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.558E-05 | 4.164E-03 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 1.558E-05 | 3.748E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.558E-05 | 3.407E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.981E-08 | 6.856E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 5.212E-08 | 5.994E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.706E-07 | 2.841E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 8.282E-07 | 4.762E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.959E-06 | 9.013E-04 |
|---|
| regulation of translation | GO:0006417 |  | 5.598E-06 | 2.146E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.371E-05 | 4.506E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.371E-05 | 3.943E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 2.052E-05 | 5.244E-03 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 2.056E-05 | 4.728E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 6.013E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.981E-08 | 6.856E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 5.212E-08 | 5.994E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.706E-07 | 2.841E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 8.282E-07 | 4.762E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.959E-06 | 9.013E-04 |
|---|
| regulation of translation | GO:0006417 |  | 5.598E-06 | 2.146E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.371E-05 | 4.506E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.371E-05 | 3.943E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 2.052E-05 | 5.244E-03 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 2.056E-05 | 4.728E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 6.013E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.981E-08 | 6.856E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 5.212E-08 | 5.994E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.706E-07 | 2.841E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 8.282E-07 | 4.762E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.959E-06 | 9.013E-04 |
|---|
| regulation of translation | GO:0006417 |  | 5.598E-06 | 2.146E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.371E-05 | 4.506E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.371E-05 | 3.943E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 2.052E-05 | 5.244E-03 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 2.056E-05 | 4.728E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 6.013E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 2.981E-08 | 6.856E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 5.212E-08 | 5.994E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.706E-07 | 2.841E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 8.282E-07 | 4.762E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.959E-06 | 9.013E-04 |
|---|
| regulation of translation | GO:0006417 |  | 5.598E-06 | 2.146E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.371E-05 | 4.506E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.371E-05 | 3.943E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 2.052E-05 | 5.244E-03 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 2.056E-05 | 4.728E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.876E-05 | 6.013E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.1E-08 | 9.729E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.555E-08 | 7.778E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.172E-07 | 2.509E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 1.323E-06 | 7.849E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 1.785E-06 | 8.47E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.166E-05 | 4.611E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.729E-05 | 5.862E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.747E-05 | 5.183E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.445E-05 | 6.446E-03 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 2.445E-05 | 5.802E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 3.258E-05 | 7.027E-03 |
|---|



