Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 2972-3-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1575 | 7.604e-03 | 9.071e-03 | 8.056e-02 |
|---|
| IPC-NIBC-129 | 0.2232 | 7.177e-02 | 9.324e-02 | 5.654e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2616 | 1.394e-02 | 2.075e-02 | 3.195e-01 |
|---|
| Loi_GPL570 | 0.2274 | 8.841e-02 | 8.787e-02 | 3.812e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1643 | 1.100e-05 | 7.100e-05 | 8.800e-05 |
|---|
| Parker_GPL1390 | 0.2318 | 1.760e-01 | 1.834e-01 | 6.945e-01 |
|---|
| Parker_GPL887 | 0.1188 | 5.835e-01 | 6.146e-01 | 6.296e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3559 | 2.086e-01 | 1.713e-01 | 7.581e-01 |
|---|
| Schmidt | 0.2378 | 1.247e-02 | 1.764e-02 | 1.548e-01 |
|---|
| Sotiriou | 0.3017 | 2.787e-03 | 8.300e-04 | 6.741e-02 |
|---|
| Wang | 0.2098 | 1.033e-01 | 7.141e-02 | 3.733e-01 |
|---|
| Zhang | 0.1982 | 6.814e-02 | 4.138e-02 | 1.699e-01 |
|---|
| Zhou | 0.4063 | 6.376e-02 | 6.768e-02 | 3.917e-01 |
|---|
Expression data for subnetwork 2972-3-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
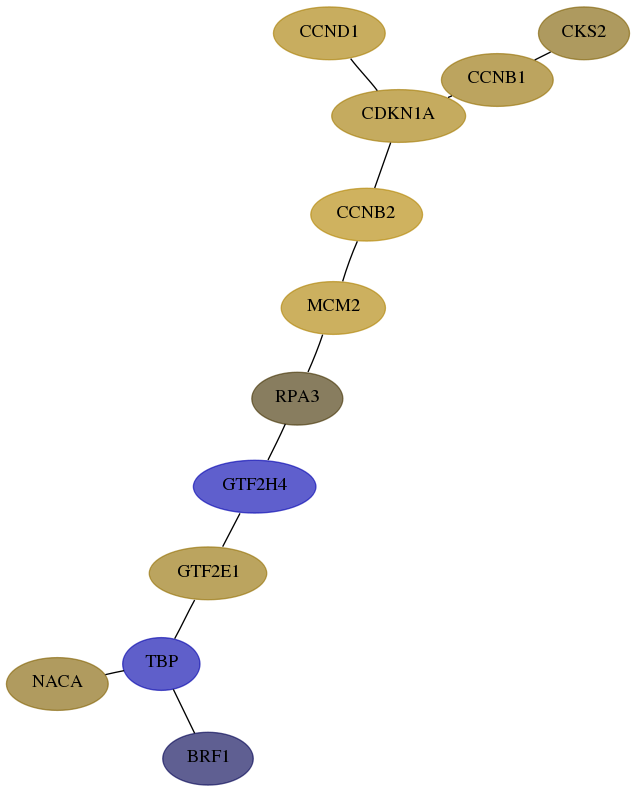
Score for each gene in subnetwork 2972-3-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| tbp |   | 2 | 104 | 322 | 308 | -0.139 | 0.211 | 0.041 | 0.154 | 0.096 | 0.049 | -0.384 | -0.102 | 0.080 | -0.033 | -0.024 | 0.061 | 0.160 |
|---|
| ccnb2 |   | 16 | 13 | 153 | 125 | 0.164 | 0.176 | 0.204 | 0.123 | 0.172 | 0.178 | 0.090 | 0.366 | 0.240 | 0.169 | 0.144 | 0.211 | 0.463 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| mcm2 |   | 4 | 60 | 216 | 192 | 0.151 | 0.254 | 0.188 | 0.128 | 0.154 | 0.220 | -0.034 | 0.267 | 0.224 | 0.200 | 0.101 | 0.012 | 0.255 |
|---|
| ccnb1 |   | 36 | 7 | 78 | 59 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | 0.152 | 0.167 | 0.398 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| cks2 |   | 44 | 6 | 73 | 54 | 0.050 | 0.233 | 0.170 | 0.103 | 0.186 | 0.172 | 0.093 | 0.167 | 0.263 | 0.151 | 0.145 | 0.284 | 0.383 |
|---|
| gtf2e1 |   | 2 | 104 | 322 | 308 | 0.080 | 0.101 | 0.138 | 0.034 | 0.106 | -0.011 | -0.129 | 0.182 | 0.073 | 0.183 | 0.060 | 0.048 | 0.085 |
|---|
| rpa3 |   | 2 | 104 | 216 | 208 | 0.011 | 0.107 | 0.109 | 0.001 | 0.162 | 0.044 | -0.259 | 0.191 | 0.126 | 0.132 | 0.149 | 0.043 | 0.108 |
|---|
| brf1 |   | 1 | 164 | 361 | 361 | -0.017 | -0.092 | 0.077 | -0.034 | 0.071 | 0.049 | 0.345 | 0.099 | 0.075 | 0.079 | 0.149 | -0.034 | 0.209 |
|---|
| gtf2h4 |   | 1 | 164 | 361 | 361 | -0.158 | 0.147 | 0.020 | 0.115 | 0.112 | 0.230 | -0.369 | 0.190 | 0.187 | 0.053 | 0.006 | -0.039 | 0.298 |
|---|
| naca |   | 4 | 60 | 120 | 108 | 0.052 | 0.141 | 0.119 | 0.132 | 0.102 | -0.017 | -0.128 | 0.081 | 0.076 | 0.100 | 0.129 | 0.147 | 0.138 |
|---|
GO Enrichment output for subnetwork 2972-3-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| transcription initiation | GO:0006352 |  | 1.426E-08 | 3.279E-05 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 1.064E-07 | 1.223E-04 |
|---|
| RNA elongation | GO:0006354 |  | 1.262E-07 | 9.674E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.487E-07 | 8.548E-05 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 5.393E-07 | 2.481E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.005E-06 | 3.852E-04 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.005E-06 | 3.302E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.237E-06 | 3.557E-04 |
|---|
| interphase | GO:0051325 |  | 1.357E-06 | 3.468E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.202E-06 | 2.116E-03 |
|---|
| response to UV | GO:0009411 |  | 1.13E-05 | 2.362E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| transcription initiation | GO:0006352 |  | 2.68E-09 | 6.548E-06 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 2.392E-08 | 2.922E-05 |
|---|
| RNA elongation | GO:0006354 |  | 3.241E-08 | 2.639E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.484E-08 | 2.128E-05 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 1.454E-07 | 7.104E-05 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 3.124E-07 | 1.272E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.136E-07 | 1.095E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.054E-07 | 1.238E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.654E-06 | 7.204E-04 |
|---|
| response to UV | GO:0009411 |  | 4.571E-06 | 1.117E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 5.095E-06 | 1.131E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| transcription initiation | GO:0006352 |  | 2.987E-09 | 7.188E-06 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 2.774E-08 | 3.337E-05 |
|---|
| RNA elongation | GO:0006354 |  | 3.779E-08 | 3.031E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.374E-08 | 2.631E-05 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 1.566E-07 | 7.537E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.472E-07 | 1.392E-04 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 3.74E-07 | 1.285E-04 |
|---|
| interphase | GO:0051325 |  | 3.935E-07 | 1.183E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.853E-07 | 1.297E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.176E-06 | 7.642E-04 |
|---|
| response to UV | GO:0009411 |  | 4.888E-06 | 1.069E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| transcription initiation | GO:0006352 |  | 2.68E-09 | 6.548E-06 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 2.392E-08 | 2.922E-05 |
|---|
| RNA elongation | GO:0006354 |  | 3.241E-08 | 2.639E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.484E-08 | 2.128E-05 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 1.454E-07 | 7.104E-05 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 3.124E-07 | 1.272E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.136E-07 | 1.095E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.054E-07 | 1.238E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.654E-06 | 7.204E-04 |
|---|
| response to UV | GO:0009411 |  | 4.571E-06 | 1.117E-03 |
|---|
| nucleotide-excision repair | GO:0006289 |  | 5.095E-06 | 1.131E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| transcription initiation | GO:0006352 |  | 2.987E-09 | 7.188E-06 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 2.774E-08 | 3.337E-05 |
|---|
| RNA elongation | GO:0006354 |  | 3.779E-08 | 3.031E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.374E-08 | 2.631E-05 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 1.566E-07 | 7.537E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.472E-07 | 1.392E-04 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 3.74E-07 | 1.285E-04 |
|---|
| interphase | GO:0051325 |  | 3.935E-07 | 1.183E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.853E-07 | 1.297E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.176E-06 | 7.642E-04 |
|---|
| response to UV | GO:0009411 |  | 4.888E-06 | 1.069E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 8.297E-11 | 2.963E-07 |
|---|
| transcription initiation | GO:0006352 |  | 3.625E-09 | 6.472E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 2.306E-08 | 2.745E-05 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 4.041E-08 | 3.607E-05 |
|---|
| RNA elongation | GO:0006354 |  | 5.24E-08 | 3.742E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.693E-08 | 3.388E-05 |
|---|
| DNA-directed RNA polymerase II. holoenzyme | GO:0016591 |  | 1.464E-07 | 7.469E-05 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 1.561E-07 | 6.968E-05 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 1.881E-07 | 7.462E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.789E-07 | 9.959E-05 |
|---|
| kinase regulator activity | GO:0019207 |  | 3.306E-07 | 1.073E-04 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 9.653E-11 | 3.522E-07 |
|---|
| transcription initiation | GO:0006352 |  | 6.985E-10 | 1.274E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.125E-08 | 1.368E-05 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 1.296E-08 | 1.182E-05 |
|---|
| RNA elongation | GO:0006354 |  | 1.701E-08 | 1.241E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.38E-08 | 1.447E-05 |
|---|
| DNA-directed RNA polymerase II. holoenzyme | GO:0016591 |  | 4.322E-08 | 2.252E-05 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 4.952E-08 | 2.258E-05 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 6.828E-08 | 2.768E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.122E-07 | 4.092E-05 |
|---|
| kinase regulator activity | GO:0019207 |  | 1.211E-07 | 4.017E-05 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| transcription initiation | GO:0006352 |  | 2.987E-09 | 7.188E-06 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 2.774E-08 | 3.337E-05 |
|---|
| RNA elongation | GO:0006354 |  | 3.779E-08 | 3.031E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.374E-08 | 2.631E-05 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 1.566E-07 | 7.537E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.472E-07 | 1.392E-04 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 3.74E-07 | 1.285E-04 |
|---|
| interphase | GO:0051325 |  | 3.935E-07 | 1.183E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.853E-07 | 1.297E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.176E-06 | 7.642E-04 |
|---|
| response to UV | GO:0009411 |  | 4.888E-06 | 1.069E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| transcription initiation | GO:0006352 |  | 1.426E-08 | 3.279E-05 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 1.064E-07 | 1.223E-04 |
|---|
| RNA elongation | GO:0006354 |  | 1.262E-07 | 9.674E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.487E-07 | 8.548E-05 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 5.393E-07 | 2.481E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.005E-06 | 3.852E-04 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.005E-06 | 3.302E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.237E-06 | 3.557E-04 |
|---|
| interphase | GO:0051325 |  | 1.357E-06 | 3.468E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.202E-06 | 2.116E-03 |
|---|
| response to UV | GO:0009411 |  | 1.13E-05 | 2.362E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| transcription initiation | GO:0006352 |  | 1.426E-08 | 3.279E-05 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 1.064E-07 | 1.223E-04 |
|---|
| RNA elongation | GO:0006354 |  | 1.262E-07 | 9.674E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.487E-07 | 8.548E-05 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 5.393E-07 | 2.481E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.005E-06 | 3.852E-04 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.005E-06 | 3.302E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.237E-06 | 3.557E-04 |
|---|
| interphase | GO:0051325 |  | 1.357E-06 | 3.468E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.202E-06 | 2.116E-03 |
|---|
| response to UV | GO:0009411 |  | 1.13E-05 | 2.362E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| transcription initiation | GO:0006352 |  | 1.426E-08 | 3.279E-05 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 1.064E-07 | 1.223E-04 |
|---|
| RNA elongation | GO:0006354 |  | 1.262E-07 | 9.674E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.487E-07 | 8.548E-05 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 5.393E-07 | 2.481E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.005E-06 | 3.852E-04 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.005E-06 | 3.302E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.237E-06 | 3.557E-04 |
|---|
| interphase | GO:0051325 |  | 1.357E-06 | 3.468E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.202E-06 | 2.116E-03 |
|---|
| response to UV | GO:0009411 |  | 1.13E-05 | 2.362E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| transcription initiation | GO:0006352 |  | 1.426E-08 | 3.279E-05 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 1.064E-07 | 1.223E-04 |
|---|
| RNA elongation | GO:0006354 |  | 1.262E-07 | 9.674E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.487E-07 | 8.548E-05 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 5.393E-07 | 2.481E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.005E-06 | 3.852E-04 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 1.005E-06 | 3.302E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.237E-06 | 3.557E-04 |
|---|
| interphase | GO:0051325 |  | 1.357E-06 | 3.468E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.202E-06 | 2.116E-03 |
|---|
| response to UV | GO:0009411 |  | 1.13E-05 | 2.362E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| transcription initiation | GO:0006352 |  | 7.123E-09 | 1.69E-05 |
|---|
| RNA elongation from RNA polymerase II promoter | GO:0006368 |  | 5.79E-08 | 6.87E-05 |
|---|
| RNA elongation | GO:0006354 |  | 6.823E-08 | 5.397E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.987E-08 | 4.738E-05 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 3.107E-07 | 1.474E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 5.632E-07 | 2.227E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.972E-07 | 2.025E-04 |
|---|
| nucleotide-excision repair. DNA damage removal | GO:0000718 |  | 6.518E-07 | 1.933E-04 |
|---|
| interphase | GO:0051325 |  | 6.545E-07 | 1.726E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.545E-06 | 1.316E-03 |
|---|
| response to UV | GO:0009411 |  | 6.779E-06 | 1.462E-03 |
|---|



