Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 2810-3-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2248 | 6.346e-03 | 7.559e-03 | 7.015e-02 |
|---|
| IPC-NIBC-129 | 0.3145 | 3.995e-02 | 5.012e-02 | 3.878e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2703 | 5.280e-04 | 1.093e-03 | 3.317e-02 |
|---|
| Loi_GPL570 | 0.3789 | 2.521e-02 | 2.498e-02 | 1.504e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1458 | 9.000e-06 | 5.700e-05 | 6.100e-05 |
|---|
| Parker_GPL1390 | 0.2745 | 6.936e-02 | 6.689e-02 | 4.499e-01 |
|---|
| Parker_GPL887 | 0.3078 | 1.757e-01 | 2.039e-01 | 1.632e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.4101 | 4.792e-01 | 4.455e-01 | 9.623e-01 |
|---|
| Schmidt | 0.2412 | 6.984e-03 | 1.013e-02 | 9.859e-02 |
|---|
| Sotiriou | 0.3370 | 2.840e-03 | 8.500e-04 | 6.832e-02 |
|---|
| Wang | 0.1885 | 1.389e-01 | 1.010e-01 | 4.503e-01 |
|---|
| Zhang | 0.1096 | 9.268e-02 | 5.980e-02 | 2.246e-01 |
|---|
| Zhou | 0.4052 | 3.788e-02 | 3.817e-02 | 2.442e-01 |
|---|
Expression data for subnetwork 2810-3-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
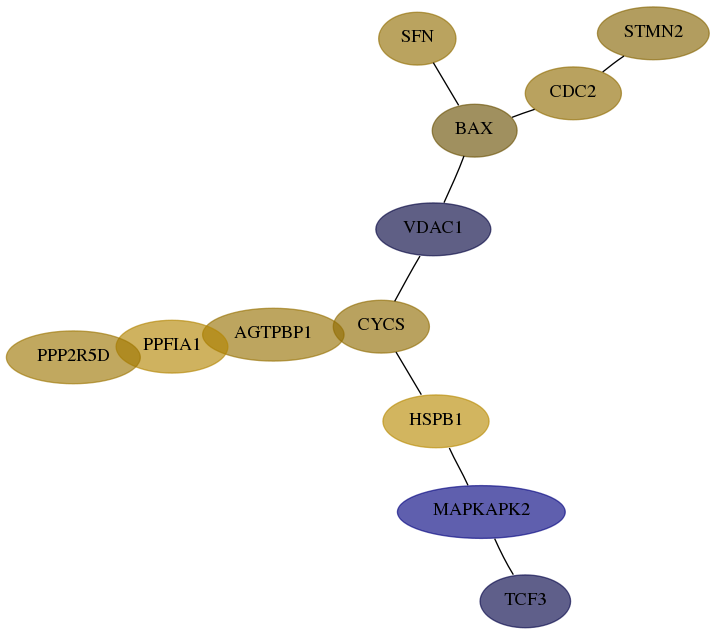
Score for each gene in subnetwork 2810-3-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| ppp2r5d |   | 3 | 79 | 17 | 22 | 0.098 | 0.040 | 0.057 | 0.270 | 0.092 | 0.237 | 0.299 | 0.241 | 0.017 | 0.103 | 0.033 | 0.048 | 0.162 |
|---|
| agtpbp1 |   | 11 | 21 | 17 | 13 | 0.086 | 0.219 | 0.009 | 0.181 | 0.071 | 0.078 | 0.296 | 0.026 | 0.110 | 0.151 | 0.103 | 0.064 | 0.226 |
|---|
| stmn2 |   | 29 | 8 | 1 | 3 | 0.057 | 0.106 | 0.005 | 0.052 | 0.110 | 0.071 | -0.099 | 0.006 | 0.034 | 0.103 | 0.075 | 0.113 | -0.039 |
|---|
| cycs |   | 10 | 25 | 17 | 14 | 0.073 | 0.206 | 0.080 | 0.094 | 0.117 | -0.056 | 0.202 | 0.272 | 0.157 | 0.077 | 0.190 | 0.135 | 0.234 |
|---|
| bax |   | 9 | 27 | 17 | 15 | 0.029 | 0.185 | 0.233 | 0.192 | 0.088 | -0.043 | 0.184 | 0.172 | 0.175 | 0.019 | -0.094 | 0.041 | 0.235 |
|---|
| vdac1 |   | 1 | 164 | 29 | 56 | -0.010 | 0.132 | 0.084 | 0.199 | 0.145 | 0.005 | 0.037 | 0.273 | -0.097 | 0.171 | -0.047 | -0.063 | 0.144 |
|---|
| ppfia1 |   | 8 | 33 | 17 | 16 | 0.164 | 0.080 | 0.149 | 0.269 | 0.151 | 0.214 | 0.093 | 0.225 | 0.036 | 0.176 | 0.153 | 0.127 | 0.272 |
|---|
| tcf3 |   | 3 | 79 | 17 | 22 | -0.012 | 0.282 | 0.055 | 0.045 | 0.037 | 0.176 | 0.318 | 0.006 | 0.094 | -0.056 | 0.040 | 0.001 | 0.167 |
|---|
| hspb1 |   | 19 | 11 | 17 | 11 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | -0.026 | -0.065 | 0.289 | -0.021 | 0.201 | 0.125 | -0.077 | 0.290 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| mapkapk2 |   | 13 | 19 | 17 | 12 | -0.050 | 0.102 | 0.156 | -0.116 | 0.090 | 0.168 | 0.303 | 0.074 | 0.171 | 0.117 | 0.085 | -0.124 | 0.312 |
|---|
| sfn |   | 50 | 4 | 29 | 17 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.025 | 0.079 | 0.181 |
|---|
GO Enrichment output for subnetwork 2810-3-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| B cell lineage commitment | GO:0002326 |  | 1.145E-07 | 2.634E-04 |
|---|
| regulation of caspase activity | GO:0043281 |  | 2.864E-06 | 3.294E-03 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 3.236E-06 | 2.481E-03 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 4.32E-06 | 2.484E-03 |
|---|
| B cell differentiation | GO:0030183 |  | 3.125E-05 | 0.01437287 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 3.433E-05 | 0.01315802 |
|---|
| activation of caspase activity | GO:0006919 |  | 3.891E-05 | 0.01278403 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 4.801E-05 | 0.01380275 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 5.425E-05 | 0.01386384 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 6.395E-05 | 0.01470891 |
|---|
| B cell activation | GO:0042113 |  | 1.346E-04 | 0.02814964 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of caspase activity | GO:0043281 |  | 3.893E-09 | 9.511E-06 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 4.767E-09 | 5.823E-06 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 6.565E-09 | 5.346E-06 |
|---|
| B cell lineage commitment | GO:0002326 |  | 1.705E-08 | 1.041E-05 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.727E-08 | 1.332E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 8.016E-08 | 3.264E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.369E-07 | 4.779E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 3.295E-07 | 1.006E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 6.43E-07 | 1.745E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 7.441E-07 | 1.818E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 9.769E-07 | 2.17E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of caspase activity | GO:0043281 |  | 9.222E-09 | 2.219E-05 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 1.06E-08 | 1.276E-05 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 1.478E-08 | 1.186E-05 |
|---|
| B cell lineage commitment | GO:0002326 |  | 3.113E-08 | 1.872E-05 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 4.978E-08 | 2.395E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.509E-07 | 6.052E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 2.633E-07 | 9.049E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 6.008E-07 | 1.807E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.172E-06 | 3.133E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.356E-06 | 3.263E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 1.78E-06 | 3.893E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of caspase activity | GO:0043281 |  | 3.893E-09 | 9.511E-06 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 4.767E-09 | 5.823E-06 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 6.565E-09 | 5.346E-06 |
|---|
| B cell lineage commitment | GO:0002326 |  | 1.705E-08 | 1.041E-05 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 2.727E-08 | 1.332E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 8.016E-08 | 3.264E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 1.369E-07 | 4.779E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 3.295E-07 | 1.006E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 6.43E-07 | 1.745E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 7.441E-07 | 1.818E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 9.769E-07 | 2.17E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of caspase activity | GO:0043281 |  | 9.222E-09 | 2.219E-05 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 1.06E-08 | 1.276E-05 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 1.478E-08 | 1.186E-05 |
|---|
| B cell lineage commitment | GO:0002326 |  | 3.113E-08 | 1.872E-05 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 4.978E-08 | 2.395E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.509E-07 | 6.052E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 2.633E-07 | 9.049E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 6.008E-07 | 1.807E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.172E-06 | 3.133E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.356E-06 | 3.263E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 1.78E-06 | 3.893E-04 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of caspase activity | GO:0043281 |  | 1.258E-07 | 4.493E-04 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 1.418E-07 | 2.532E-04 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 1.884E-07 | 2.242E-04 |
|---|
| protein phosphatase type 2A complex | GO:0000159 |  | 2.511E-07 | 2.241E-04 |
|---|
| protein serine/threonine phosphatase complex | GO:0008287 |  | 2E-06 | 1.428E-03 |
|---|
| activation of caspase activity | GO:0006919 |  | 4.618E-06 | 2.748E-03 |
|---|
| bHLH transcription factor binding | GO:0043425 |  | 5.061E-06 | 2.582E-03 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 6.706E-06 | 2.993E-03 |
|---|
| B cell lineage commitment | GO:0002326 |  | 7.589E-06 | 3.011E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.062E-05 | 3.792E-03 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 1.062E-05 | 3.448E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.454E-08 | 5.304E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 1.267E-07 | 2.312E-04 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.507E-07 | 1.833E-04 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 1.546E-07 | 1.41E-04 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 1.986E-07 | 1.449E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.316E-07 | 1.408E-04 |
|---|
| protein phosphatase type 2A complex | GO:0000159 |  | 2.811E-07 | 1.465E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 4.706E-07 | 2.146E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 5.488E-07 | 2.224E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 5.488E-07 | 2.002E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 6.351E-07 | 2.106E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of caspase activity | GO:0043281 |  | 9.222E-09 | 2.219E-05 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 1.06E-08 | 1.276E-05 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 1.478E-08 | 1.186E-05 |
|---|
| B cell lineage commitment | GO:0002326 |  | 3.113E-08 | 1.872E-05 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 4.978E-08 | 2.395E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.509E-07 | 6.052E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 2.633E-07 | 9.049E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 6.008E-07 | 1.807E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.172E-06 | 3.133E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.356E-06 | 3.263E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 1.78E-06 | 3.893E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| B cell lineage commitment | GO:0002326 |  | 1.145E-07 | 2.634E-04 |
|---|
| regulation of caspase activity | GO:0043281 |  | 2.864E-06 | 3.294E-03 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 3.236E-06 | 2.481E-03 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 4.32E-06 | 2.484E-03 |
|---|
| B cell differentiation | GO:0030183 |  | 3.125E-05 | 0.01437287 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 3.433E-05 | 0.01315802 |
|---|
| activation of caspase activity | GO:0006919 |  | 3.891E-05 | 0.01278403 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 4.801E-05 | 0.01380275 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 5.425E-05 | 0.01386384 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 6.395E-05 | 0.01470891 |
|---|
| B cell activation | GO:0042113 |  | 1.346E-04 | 0.02814964 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| B cell lineage commitment | GO:0002326 |  | 1.145E-07 | 2.634E-04 |
|---|
| regulation of caspase activity | GO:0043281 |  | 2.864E-06 | 3.294E-03 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 3.236E-06 | 2.481E-03 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 4.32E-06 | 2.484E-03 |
|---|
| B cell differentiation | GO:0030183 |  | 3.125E-05 | 0.01437287 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 3.433E-05 | 0.01315802 |
|---|
| activation of caspase activity | GO:0006919 |  | 3.891E-05 | 0.01278403 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 4.801E-05 | 0.01380275 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 5.425E-05 | 0.01386384 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 6.395E-05 | 0.01470891 |
|---|
| B cell activation | GO:0042113 |  | 1.346E-04 | 0.02814964 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| B cell lineage commitment | GO:0002326 |  | 1.145E-07 | 2.634E-04 |
|---|
| regulation of caspase activity | GO:0043281 |  | 2.864E-06 | 3.294E-03 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 3.236E-06 | 2.481E-03 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 4.32E-06 | 2.484E-03 |
|---|
| B cell differentiation | GO:0030183 |  | 3.125E-05 | 0.01437287 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 3.433E-05 | 0.01315802 |
|---|
| activation of caspase activity | GO:0006919 |  | 3.891E-05 | 0.01278403 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 4.801E-05 | 0.01380275 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 5.425E-05 | 0.01386384 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 6.395E-05 | 0.01470891 |
|---|
| B cell activation | GO:0042113 |  | 1.346E-04 | 0.02814964 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| B cell lineage commitment | GO:0002326 |  | 1.145E-07 | 2.634E-04 |
|---|
| regulation of caspase activity | GO:0043281 |  | 2.864E-06 | 3.294E-03 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 3.236E-06 | 2.481E-03 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 4.32E-06 | 2.484E-03 |
|---|
| B cell differentiation | GO:0030183 |  | 3.125E-05 | 0.01437287 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 3.433E-05 | 0.01315802 |
|---|
| activation of caspase activity | GO:0006919 |  | 3.891E-05 | 0.01278403 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 4.801E-05 | 0.01380275 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 5.425E-05 | 0.01386384 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 6.395E-05 | 0.01470891 |
|---|
| B cell activation | GO:0042113 |  | 1.346E-04 | 0.02814964 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| B cell lineage commitment | GO:0002326 |  | 1.195E-07 | 2.836E-04 |
|---|
| regulation of caspase activity | GO:0043281 |  | 3.668E-06 | 4.352E-03 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 4.121E-06 | 3.26E-03 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 5.433E-06 | 3.223E-03 |
|---|
| B cell differentiation | GO:0030183 |  | 3.258E-05 | 0.0154628 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 3.513E-05 | 0.01389539 |
|---|
| regulation of protein homodimerization activity | GO:0043496 |  | 4.914E-05 | 0.01665826 |
|---|
| activation of caspase activity | GO:0006919 |  | 4.974E-05 | 0.01475479 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 6.546E-05 | 0.01725845 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 6.788E-05 | 0.01610813 |
|---|
| B cell activation | GO:0042113 |  | 1.468E-04 | 0.03167246 |
|---|



