Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 27158-3-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2009 | 8.655e-03 | 1.034e-02 | 8.888e-02 |
|---|
| IPC-NIBC-129 | 0.2076 | 8.127e-02 | 1.062e-01 | 6.058e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2553 | 2.717e-03 | 4.769e-03 | 1.127e-01 |
|---|
| Loi_GPL570 | 0.3036 | 1.088e-01 | 1.082e-01 | 4.368e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1869 | 1.440e-04 | 6.840e-04 | 3.513e-03 |
|---|
| Parker_GPL1390 | 0.2227 | 9.707e-02 | 9.650e-02 | 5.331e-01 |
|---|
| Parker_GPL887 | 0.0817 | 7.171e-01 | 7.416e-01 | 7.728e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3675 | 1.583e-01 | 1.246e-01 | 6.681e-01 |
|---|
| Schmidt | 0.2003 | 8.986e-03 | 1.290e-02 | 1.203e-01 |
|---|
| Sotiriou | 0.3217 | 1.524e-03 | 3.870e-04 | 4.337e-02 |
|---|
| Wang | 0.2069 | 7.167e-02 | 4.661e-02 | 2.927e-01 |
|---|
| Zhang | 0.2259 | 7.102e-02 | 4.348e-02 | 1.764e-01 |
|---|
| Zhou | 0.4426 | 5.710e-02 | 5.998e-02 | 3.566e-01 |
|---|
Expression data for subnetwork 27158-3-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
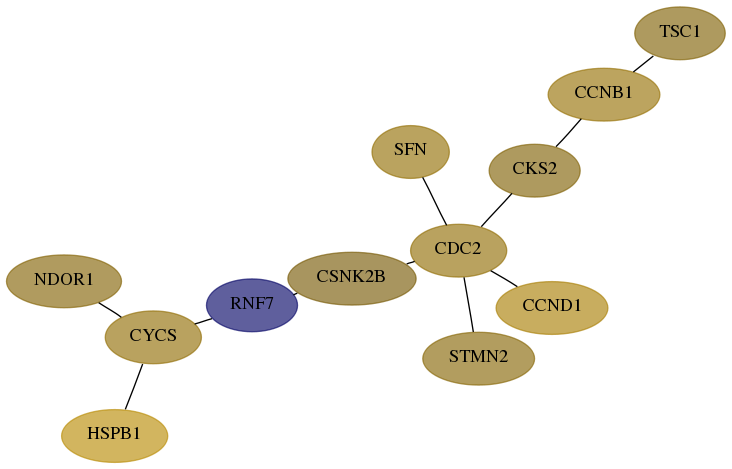
Score for each gene in subnetwork 27158-3-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| rnf7 |   | 1 | 164 | 225 | 233 | -0.027 | -0.106 | 0.061 | 0.093 | 0.074 | 0.107 | -0.535 | 0.162 | 0.083 | 0.035 | 0.059 | 0.257 | 0.288 |
|---|
| stmn2 |   | 29 | 8 | 1 | 3 | 0.057 | 0.106 | 0.005 | 0.052 | 0.110 | 0.071 | -0.099 | 0.006 | 0.034 | 0.103 | 0.075 | 0.113 | -0.039 |
|---|
| cycs |   | 10 | 25 | 17 | 14 | 0.073 | 0.206 | 0.080 | 0.094 | 0.117 | -0.056 | 0.202 | 0.272 | 0.157 | 0.077 | 0.190 | 0.135 | 0.234 |
|---|
| ccnb1 |   | 36 | 7 | 78 | 59 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | 0.152 | 0.167 | 0.398 |
|---|
| ndor1 |   | 1 | 164 | 225 | 233 | 0.052 | 0.053 | 0.101 | 0.023 | 0.065 | 0.042 | 0.070 | -0.010 | 0.095 | 0.139 | 0.001 | 0.021 | -0.060 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| cks2 |   | 44 | 6 | 73 | 54 | 0.050 | 0.233 | 0.170 | 0.103 | 0.186 | 0.172 | 0.093 | 0.167 | 0.263 | 0.151 | 0.145 | 0.284 | 0.383 |
|---|
| hspb1 |   | 19 | 11 | 17 | 11 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | -0.026 | -0.065 | 0.289 | -0.021 | 0.201 | 0.125 | -0.077 | 0.290 |
|---|
| tsc1 |   | 47 | 5 | 17 | 8 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.043 | -0.007 | 0.207 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| csnk2b |   | 1 | 164 | 225 | 233 | 0.039 | 0.147 | 0.116 | 0.078 | 0.110 | 0.233 | -0.494 | 0.221 | 0.113 | 0.028 | 0.009 | -0.102 | 0.136 |
|---|
| sfn |   | 50 | 4 | 29 | 17 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.025 | 0.079 | 0.181 |
|---|
GO Enrichment output for subnetwork 27158-3-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.563E-08 | 1.969E-04 |
|---|
| response to unfolded protein | GO:0006986 |  | 4.341E-07 | 4.992E-04 |
|---|
| response to protein stimulus | GO:0051789 |  | 5.615E-07 | 4.305E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.954E-06 | 2.848E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.861E-06 | 3.156E-03 |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 6.861E-06 | 2.63E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 6.861E-06 | 2.254E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.029E-05 | 2.958E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 3.679E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.439E-05 | 3.311E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.785E-05 | 3.733E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to redox state | GO:0051775 |  | 1.776E-09 | 4.338E-06 |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 6.211E-09 | 7.587E-06 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 9.934E-09 | 8.089E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.32E-08 | 8.063E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.484E-08 | 1.702E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 1.008E-07 | 4.105E-05 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 1.184E-07 | 4.134E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.202E-07 | 3.671E-05 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 1.528E-07 | 4.148E-05 |
|---|
| response to unfolded protein | GO:0006986 |  | 2.225E-07 | 5.437E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.348E-07 | 5.214E-05 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to redox state | GO:0051775 |  | 2.919E-09 | 7.023E-06 |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 1.021E-08 | 1.228E-05 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.632E-08 | 1.309E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.554E-08 | 1.536E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.735E-08 | 3.241E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 1.742E-07 | 6.986E-05 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 1.947E-07 | 6.691E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.974E-07 | 5.937E-05 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 2.536E-07 | 6.781E-05 |
|---|
| response to unfolded protein | GO:0006986 |  | 3.409E-07 | 8.201E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 3.854E-07 | 8.431E-05 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to redox state | GO:0051775 |  | 1.776E-09 | 4.338E-06 |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 6.211E-09 | 7.587E-06 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 9.934E-09 | 8.089E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.32E-08 | 8.063E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.484E-08 | 1.702E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 1.008E-07 | 4.105E-05 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 1.184E-07 | 4.134E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.202E-07 | 3.671E-05 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 1.528E-07 | 4.148E-05 |
|---|
| response to unfolded protein | GO:0006986 |  | 2.225E-07 | 5.437E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.348E-07 | 5.214E-05 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to redox state | GO:0051775 |  | 2.919E-09 | 7.023E-06 |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 1.021E-08 | 1.228E-05 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.632E-08 | 1.309E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.554E-08 | 1.536E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.735E-08 | 3.241E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 1.742E-07 | 6.986E-05 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 1.947E-07 | 6.691E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.974E-07 | 5.937E-05 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 2.536E-07 | 6.781E-05 |
|---|
| response to unfolded protein | GO:0006986 |  | 3.409E-07 | 8.201E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 3.854E-07 | 8.431E-05 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein kinase regulator activity | GO:0019887 |  | 5.971E-10 | 2.132E-06 |
|---|
| kinase regulator activity | GO:0019207 |  | 1.54E-09 | 2.75E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.805E-08 | 4.529E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 5.091E-08 | 4.545E-05 |
|---|
| response to unfolded protein | GO:0006986 |  | 1.183E-07 | 8.452E-05 |
|---|
| response to protein stimulus | GO:0051789 |  | 1.685E-07 | 1.003E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.759E-06 | 1.917E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.061E-06 | 2.259E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.589E-06 | 3.011E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.062E-05 | 3.792E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 1.062E-05 | 3.448E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.43E-08 | 1.251E-04 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 9.481E-08 | 1.729E-04 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 9.836E-08 | 1.196E-04 |
|---|
| kinase regulator activity | GO:0019207 |  | 1.744E-07 | 1.591E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.747E-06 | 2.004E-03 |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 4.447E-06 | 2.704E-03 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 4.447E-06 | 2.317E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 4.447E-06 | 2.028E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.667E-06 | 2.702E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 6.667E-06 | 2.432E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.667E-06 | 2.211E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to redox state | GO:0051775 |  | 2.919E-09 | 7.023E-06 |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 1.021E-08 | 1.228E-05 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.632E-08 | 1.309E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.554E-08 | 1.536E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.735E-08 | 3.241E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 1.742E-07 | 6.986E-05 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 1.947E-07 | 6.691E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.974E-07 | 5.937E-05 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 2.536E-07 | 6.781E-05 |
|---|
| response to unfolded protein | GO:0006986 |  | 3.409E-07 | 8.201E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 3.854E-07 | 8.431E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.563E-08 | 1.969E-04 |
|---|
| response to unfolded protein | GO:0006986 |  | 4.341E-07 | 4.992E-04 |
|---|
| response to protein stimulus | GO:0051789 |  | 5.615E-07 | 4.305E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.954E-06 | 2.848E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.861E-06 | 3.156E-03 |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 6.861E-06 | 2.63E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 6.861E-06 | 2.254E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.029E-05 | 2.958E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 3.679E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.439E-05 | 3.311E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.785E-05 | 3.733E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.563E-08 | 1.969E-04 |
|---|
| response to unfolded protein | GO:0006986 |  | 4.341E-07 | 4.992E-04 |
|---|
| response to protein stimulus | GO:0051789 |  | 5.615E-07 | 4.305E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.954E-06 | 2.848E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.861E-06 | 3.156E-03 |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 6.861E-06 | 2.63E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 6.861E-06 | 2.254E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.029E-05 | 2.958E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 3.679E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.439E-05 | 3.311E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.785E-05 | 3.733E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.563E-08 | 1.969E-04 |
|---|
| response to unfolded protein | GO:0006986 |  | 4.341E-07 | 4.992E-04 |
|---|
| response to protein stimulus | GO:0051789 |  | 5.615E-07 | 4.305E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.954E-06 | 2.848E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.861E-06 | 3.156E-03 |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 6.861E-06 | 2.63E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 6.861E-06 | 2.254E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.029E-05 | 2.958E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 3.679E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.439E-05 | 3.311E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.785E-05 | 3.733E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.563E-08 | 1.969E-04 |
|---|
| response to unfolded protein | GO:0006986 |  | 4.341E-07 | 4.992E-04 |
|---|
| response to protein stimulus | GO:0051789 |  | 5.615E-07 | 4.305E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.954E-06 | 2.848E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.861E-06 | 3.156E-03 |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 6.861E-06 | 2.63E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 6.861E-06 | 2.254E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.029E-05 | 2.958E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.439E-05 | 3.679E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.439E-05 | 3.311E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.785E-05 | 3.733E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.338E-07 | 3.174E-04 |
|---|
| response to unfolded protein | GO:0006986 |  | 7.115E-07 | 8.442E-04 |
|---|
| response to protein stimulus | GO:0051789 |  | 9.083E-07 | 7.185E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.99E-06 | 4.147E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.724E-06 | 3.666E-03 |
|---|
| induction of apoptosis by oxidative stress | GO:0008631 |  | 7.724E-06 | 3.055E-03 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 1.158E-05 | 3.926E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.158E-05 | 3.435E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.62E-05 | 4.272E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 2.159E-05 | 5.124E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.159E-05 | 4.658E-03 |
|---|



