Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 23649-2-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1329 | 8.959e-03 | 1.070e-02 | 9.123e-02 |
|---|
| IPC-NIBC-129 | 0.2634 | 3.501e-02 | 4.351e-02 | 3.524e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2536 | 1.185e-02 | 1.793e-02 | 2.910e-01 |
|---|
| Loi_GPL570 | 0.2350 | 5.121e-02 | 5.084e-02 | 2.594e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1569 | 1.000e-06 | 1.100e-05 | 3.000e-06 |
|---|
| Parker_GPL1390 | 0.2841 | 2.444e-01 | 2.600e-01 | 7.846e-01 |
|---|
| Parker_GPL887 | 0.0142 | 9.698e-01 | 9.734e-01 | 9.875e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3228 | 1.215e-01 | 9.163e-02 | 5.806e-01 |
|---|
| Schmidt | 0.2680 | 1.039e-02 | 1.482e-02 | 1.347e-01 |
|---|
| Sotiriou | 0.3128 | 1.635e-03 | 4.230e-04 | 4.569e-02 |
|---|
| Wang | 0.2141 | 1.164e-01 | 8.210e-02 | 4.030e-01 |
|---|
| Zhang | 0.2522 | 6.404e-02 | 3.841e-02 | 1.604e-01 |
|---|
| Zhou | 0.4384 | 8.719e-02 | 9.518e-02 | 5.006e-01 |
|---|
Expression data for subnetwork 23649-2-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
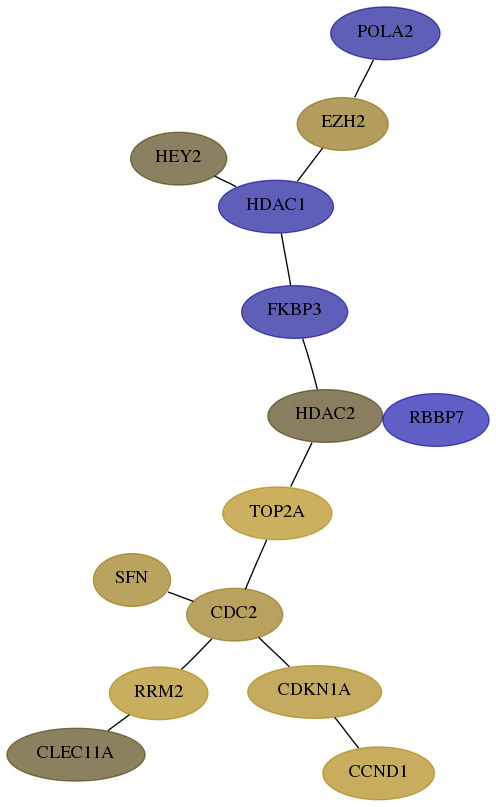
Score for each gene in subnetwork 23649-2-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| hdac1 |   | 2 | 104 | 219 | 213 | -0.076 | 0.003 | -0.041 | 0.108 | 0.111 | 0.128 | 0.164 | -0.034 | -0.074 | 0.152 | -0.019 | 0.086 | 0.292 |
|---|
| pola2 |   | 1 | 164 | 328 | 333 | -0.067 | 0.193 | 0.089 | -0.124 | 0.103 | 0.131 | 0.345 | 0.039 | 0.087 | 0.161 | 0.104 | 0.011 | 0.019 |
|---|
| rrm2 |   | 21 | 10 | 44 | 26 | 0.131 | 0.207 | 0.227 | 0.187 | 0.161 | 0.178 | 0.137 | 0.303 | 0.190 | 0.219 | 0.117 | 0.129 | 0.302 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| hey2 |   | 2 | 104 | 219 | 213 | 0.013 | 0.189 | -0.017 | 0.127 | 0.076 | 0.165 | -0.287 | -0.039 | 0.152 | 0.076 | 0.057 | -0.043 | 0.367 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| fkbp3 |   | 2 | 104 | 279 | 265 | -0.072 | 0.129 | 0.144 | 0.166 | 0.136 | -0.034 | -0.215 | 0.010 | 0.159 | 0.052 | 0.058 | 0.199 | 0.288 |
|---|
| rbbp7 |   | 3 | 79 | 219 | 206 | -0.113 | 0.089 | 0.051 | 0.076 | 0.106 | -0.053 | 0.092 | -0.020 | 0.126 | 0.138 | 0.115 | 0.168 | 0.280 |
|---|
| hdac2 |   | 2 | 104 | 279 | 265 | 0.012 | 0.245 | 0.030 | 0.077 | 0.070 | 0.307 | -0.204 | 0.183 | 0.222 | -0.058 | 0.086 | 0.133 | 0.181 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| sfn |   | 50 | 4 | 29 | 17 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.025 | 0.079 | 0.181 |
|---|
| top2a |   | 11 | 21 | 93 | 76 | 0.147 | 0.183 | 0.185 | 0.100 | 0.206 | 0.014 | 0.035 | 0.309 | 0.261 | 0.183 | 0.072 | 0.234 | 0.273 |
|---|
| clec11a |   | 17 | 12 | 44 | 27 | 0.013 | 0.101 | 0.071 | -0.085 | 0.066 | 0.082 | 0.209 | -0.126 | -0.034 | -0.001 | 0.191 | 0.059 | -0.075 |
|---|
| ezh2 |   | 2 | 104 | 328 | 316 | 0.058 | 0.253 | 0.191 | 0.110 | 0.106 | 0.142 | 0.054 | 0.250 | 0.218 | 0.172 | 0.123 | 0.126 | 0.374 |
|---|
GO Enrichment output for subnetwork 23649-2-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.451E-08 | 7.937E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.034E-08 | 6.939E-05 |
|---|
| histone deacetylation | GO:0016575 |  | 2.835E-07 | 2.174E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.52E-07 | 2.599E-04 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 9.584E-07 | 4.409E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.267E-06 | 8.69E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.508E-05 | 4.956E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.508E-05 | 4.336E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.658E-05 | 4.238E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.066E-05 | 4.752E-03 |
|---|
| DNA topological change | GO:0006265 |  | 2.261E-05 | 4.727E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.164E-08 | 1.506E-04 |
|---|
| histone deacetylation | GO:0016575 |  | 1.224E-07 | 1.495E-04 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 3.06E-07 | 2.492E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.767E-06 | 2.301E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.028E-06 | 1.968E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 4.364E-06 | 1.777E-03 |
|---|
| histone modification | GO:0016570 |  | 5.849E-06 | 2.041E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 6.543E-06 | 1.998E-03 |
|---|
| covalent chromatin modification | GO:0016569 |  | 6.559E-06 | 1.78E-03 |
|---|
| response to UV | GO:0009411 |  | 6.934E-06 | 1.694E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 9.157E-06 | 2.034E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.764E-08 | 4.245E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.23E-08 | 5.089E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.415E-07 | 1.135E-04 |
|---|
| histone deacetylation | GO:0016575 |  | 2.284E-07 | 1.374E-04 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 5.706E-07 | 2.746E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 7.7E-07 | 3.088E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 6.605E-06 | 2.27E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 6.605E-06 | 1.986E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.006E-06 | 1.873E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.491E-06 | 1.802E-03 |
|---|
| histone modification | GO:0016570 |  | 9.651E-06 | 2.111E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.164E-08 | 1.506E-04 |
|---|
| histone deacetylation | GO:0016575 |  | 1.224E-07 | 1.495E-04 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 3.06E-07 | 2.492E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.767E-06 | 2.301E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.028E-06 | 1.968E-03 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 4.364E-06 | 1.777E-03 |
|---|
| histone modification | GO:0016570 |  | 5.849E-06 | 2.041E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 6.543E-06 | 1.998E-03 |
|---|
| covalent chromatin modification | GO:0016569 |  | 6.559E-06 | 1.78E-03 |
|---|
| response to UV | GO:0009411 |  | 6.934E-06 | 1.694E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 9.157E-06 | 2.034E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.764E-08 | 4.245E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.23E-08 | 5.089E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.415E-07 | 1.135E-04 |
|---|
| histone deacetylation | GO:0016575 |  | 2.284E-07 | 1.374E-04 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 5.706E-07 | 2.746E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 7.7E-07 | 3.088E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 6.605E-06 | 2.27E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 6.605E-06 | 1.986E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.006E-06 | 1.873E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.491E-06 | 1.802E-03 |
|---|
| histone modification | GO:0016570 |  | 9.651E-06 | 2.111E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| histone deacetylase complex | GO:0000118 |  | 5.387E-09 | 1.924E-05 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 2.997E-08 | 5.352E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.202E-08 | 9.763E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 8.818E-08 | 7.872E-05 |
|---|
| histone deacetylation | GO:0016575 |  | 1.527E-07 | 1.09E-04 |
|---|
| chromatin remodeling complex | GO:0016585 |  | 1.614E-07 | 9.605E-05 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 1.942E-07 | 9.908E-05 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 2.247E-07 | 1.003E-04 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 3.623E-07 | 1.437E-04 |
|---|
| histone deacetylase activity | GO:0004407 |  | 4.345E-07 | 1.552E-04 |
|---|
| kinase regulator activity | GO:0019207 |  | 4.756E-07 | 1.544E-04 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| histone deacetylase complex | GO:0000118 |  | 2.898E-09 | 1.057E-05 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.861E-08 | 3.394E-05 |
|---|
| chromatin remodeling complex | GO:0016585 |  | 3.737E-08 | 4.544E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.793E-08 | 4.371E-05 |
|---|
| histone deacetylation | GO:0016575 |  | 9.481E-08 | 6.917E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.206E-07 | 7.333E-05 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 1.373E-07 | 7.158E-05 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 1.854E-07 | 8.454E-05 |
|---|
| kinase regulator activity | GO:0019207 |  | 2.434E-07 | 9.868E-05 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 2.699E-07 | 9.846E-05 |
|---|
| histone deacetylase activity | GO:0004407 |  | 3.204E-07 | 1.062E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.764E-08 | 4.245E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.23E-08 | 5.089E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.415E-07 | 1.135E-04 |
|---|
| histone deacetylation | GO:0016575 |  | 2.284E-07 | 1.374E-04 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 5.706E-07 | 2.746E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 7.7E-07 | 3.088E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 6.605E-06 | 2.27E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 6.605E-06 | 1.986E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.006E-06 | 1.873E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.491E-06 | 1.802E-03 |
|---|
| histone modification | GO:0016570 |  | 9.651E-06 | 2.111E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.451E-08 | 7.937E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.034E-08 | 6.939E-05 |
|---|
| histone deacetylation | GO:0016575 |  | 2.835E-07 | 2.174E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.52E-07 | 2.599E-04 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 9.584E-07 | 4.409E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.267E-06 | 8.69E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.508E-05 | 4.956E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.508E-05 | 4.336E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.658E-05 | 4.238E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.066E-05 | 4.752E-03 |
|---|
| DNA topological change | GO:0006265 |  | 2.261E-05 | 4.727E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.451E-08 | 7.937E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.034E-08 | 6.939E-05 |
|---|
| histone deacetylation | GO:0016575 |  | 2.835E-07 | 2.174E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.52E-07 | 2.599E-04 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 9.584E-07 | 4.409E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.267E-06 | 8.69E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.508E-05 | 4.956E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.508E-05 | 4.336E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.658E-05 | 4.238E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.066E-05 | 4.752E-03 |
|---|
| DNA topological change | GO:0006265 |  | 2.261E-05 | 4.727E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.451E-08 | 7.937E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.034E-08 | 6.939E-05 |
|---|
| histone deacetylation | GO:0016575 |  | 2.835E-07 | 2.174E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.52E-07 | 2.599E-04 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 9.584E-07 | 4.409E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.267E-06 | 8.69E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.508E-05 | 4.956E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.508E-05 | 4.336E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.658E-05 | 4.238E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.066E-05 | 4.752E-03 |
|---|
| DNA topological change | GO:0006265 |  | 2.261E-05 | 4.727E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 3.451E-08 | 7.937E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 6.034E-08 | 6.939E-05 |
|---|
| histone deacetylation | GO:0016575 |  | 2.835E-07 | 2.174E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.52E-07 | 2.599E-04 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 9.584E-07 | 4.409E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.267E-06 | 8.69E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.508E-05 | 4.956E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.508E-05 | 4.336E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.658E-05 | 4.238E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.066E-05 | 4.752E-03 |
|---|
| DNA topological change | GO:0006265 |  | 2.261E-05 | 4.727E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 5.387E-08 | 1.278E-04 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 8.612E-08 | 1.022E-04 |
|---|
| histone deacetylation | GO:0016575 |  | 3.373E-07 | 2.668E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.59E-07 | 2.723E-04 |
|---|
| protein amino acid deacetylation | GO:0006476 |  | 1.038E-06 | 4.928E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 2.342E-06 | 9.264E-04 |
|---|
| positive regulation of viral genome replication | GO:0045070 |  | 1.393E-05 | 4.721E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 1.393E-05 | 4.131E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.719E-05 | 4.531E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.98E-05 | 4.697E-03 |
|---|
| response to UV | GO:0009411 |  | 2.418E-05 | 5.216E-03 |
|---|



