Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 22907-2-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1781 | 1.077e-02 | 1.288e-02 | 1.047e-01 |
|---|
| IPC-NIBC-129 | 0.2467 | 3.640e-02 | 4.536e-02 | 3.626e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2447 | 4.399e-02 | 5.839e-02 | 5.683e-01 |
|---|
| Loi_GPL570 | 0.1728 | 6.434e-02 | 6.390e-02 | 3.060e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1442 | 2.000e-06 | 1.400e-05 | 5.000e-06 |
|---|
| Parker_GPL1390 | 0.2813 | 1.330e-01 | 1.357e-01 | 6.171e-01 |
|---|
| Parker_GPL887 | 0.1053 | 6.304e-01 | 6.595e-01 | 6.811e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3351 | 9.310e-02 | 6.731e-02 | 4.962e-01 |
|---|
| Schmidt | 0.2639 | 8.726e-03 | 1.254e-02 | 1.176e-01 |
|---|
| Sotiriou | 0.3235 | 1.873e-03 | 5.020e-04 | 5.051e-02 |
|---|
| Wang | 0.2151 | 1.425e-01 | 1.040e-01 | 4.573e-01 |
|---|
| Zhang | 0.2781 | 6.309e-02 | 3.773e-02 | 1.582e-01 |
|---|
| Zhou | 0.4302 | 7.769e-02 | 8.397e-02 | 4.590e-01 |
|---|
Expression data for subnetwork 22907-2-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
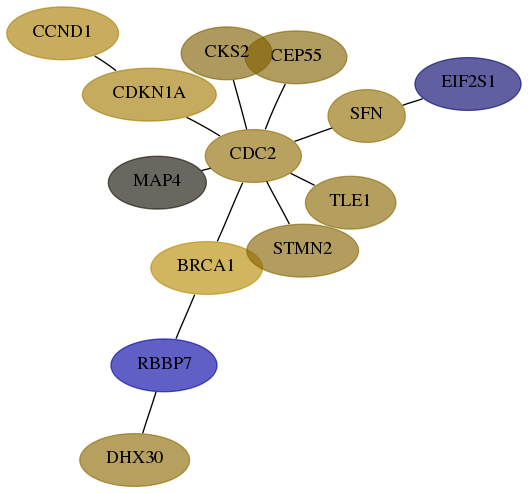
Score for each gene in subnetwork 22907-2-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| stmn2 |   | 29 | 8 | 1 | 3 | 0.057 | 0.106 | 0.005 | 0.052 | 0.110 | 0.071 | -0.099 | 0.006 | 0.034 | 0.103 | 0.075 | 0.113 | -0.039 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| tle1 |   | 23 | 9 | 78 | 60 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | -0.028 | 0.068 | 0.025 |
|---|
| cep55 |   | 3 | 79 | 141 | 137 | 0.054 | 0.210 | 0.148 | 0.129 | 0.135 | 0.162 | 0.133 | 0.209 | 0.202 | 0.138 | 0.144 | 0.115 | 0.330 |
|---|
| brca1 |   | 8 | 33 | 175 | 150 | 0.192 | 0.052 | 0.112 | 0.022 | 0.125 | -0.124 | 0.305 | 0.113 | 0.077 | 0.168 | 0.155 | 0.064 | 0.209 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| cks2 |   | 44 | 6 | 73 | 54 | 0.050 | 0.233 | 0.170 | 0.103 | 0.186 | 0.172 | 0.093 | 0.167 | 0.263 | 0.151 | 0.145 | 0.284 | 0.383 |
|---|
| rbbp7 |   | 3 | 79 | 219 | 206 | -0.113 | 0.089 | 0.051 | 0.076 | 0.106 | -0.053 | 0.092 | -0.020 | 0.126 | 0.138 | 0.115 | 0.168 | 0.280 |
|---|
| dhx30 |   | 1 | 164 | 239 | 242 | 0.066 | -0.039 | 0.122 | -0.072 | 0.099 | 0.272 | -0.261 | 0.326 | 0.065 | 0.160 | -0.028 | 0.015 | 0.304 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| map4 |   | 16 | 13 | 73 | 55 | 0.002 | 0.046 | 0.067 | -0.043 | 0.034 | 0.119 | 0.159 | -0.058 | -0.020 | 0.043 | 0.108 | -0.074 | 0.302 |
|---|
| sfn |   | 50 | 4 | 29 | 17 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.025 | 0.079 | 0.181 |
|---|
| eif2s1 |   | 14 | 17 | 73 | 57 | -0.031 | 0.151 | 0.253 | 0.182 | 0.098 | 0.167 | 0.005 | 0.270 | 0.233 | 0.038 | 0.122 | 0.239 | 0.265 |
|---|
GO Enrichment output for subnetwork 22907-2-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.614E-08 | 3.712E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.623E-07 | 5.317E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 6.378E-06 | 4.89E-03 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 9.334E-06 | 5.367E-03 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 1.451E-05 | 6.676E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 2.466E-05 | 9.455E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 3.696E-05 | 0.01214401 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 3.696E-05 | 0.01062601 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 3.696E-05 | 9.445E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.35E-05 | 0.01000551 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 5.169E-05 | 0.01080849 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.379E-09 | 5.813E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.011E-07 | 1.235E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 1.813E-06 | 1.476E-03 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 3.529E-06 | 2.155E-03 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 6.076E-06 | 2.969E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 8.865E-06 | 3.61E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.18E-05 | 4.119E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 1.329E-05 | 4.058E-03 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 1.329E-05 | 3.607E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 1.859E-05 | 4.543E-03 |
|---|
| response to UV | GO:0009411 |  | 2.028E-05 | 4.503E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.414E-09 | 1.303E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.951E-07 | 2.347E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 2.972E-06 | 2.384E-03 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 4.708E-06 | 2.832E-03 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 8.393E-06 | 4.039E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.234E-05 | 4.95E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 1.85E-05 | 6.36E-03 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 1.85E-05 | 5.565E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.93E-05 | 5.16E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 2.589E-05 | 6.228E-03 |
|---|
| response to UV | GO:0009411 |  | 2.963E-05 | 6.48E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.379E-09 | 5.813E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.011E-07 | 1.235E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 1.813E-06 | 1.476E-03 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 3.529E-06 | 2.155E-03 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 6.076E-06 | 2.969E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 8.865E-06 | 3.61E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.18E-05 | 4.119E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 1.329E-05 | 4.058E-03 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 1.329E-05 | 3.607E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 1.859E-05 | 4.543E-03 |
|---|
| response to UV | GO:0009411 |  | 2.028E-05 | 4.503E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.414E-09 | 1.303E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.951E-07 | 2.347E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 2.972E-06 | 2.384E-03 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 4.708E-06 | 2.832E-03 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 8.393E-06 | 4.039E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.234E-05 | 4.95E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 1.85E-05 | 6.36E-03 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 1.85E-05 | 5.565E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.93E-05 | 5.16E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 2.589E-05 | 6.228E-03 |
|---|
| response to UV | GO:0009411 |  | 2.963E-05 | 6.48E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 8.297E-11 | 2.963E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.851E-10 | 5.09E-07 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 1.02E-09 | 1.214E-06 |
|---|
| kinase regulator activity | GO:0019207 |  | 2.63E-09 | 2.348E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 2.306E-08 | 1.647E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.06E-08 | 1.821E-05 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 1.495E-07 | 7.626E-05 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 8.274E-07 | 3.693E-04 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 9.398E-07 | 3.729E-04 |
|---|
| protein kinase inhibitor activity | GO:0004860 |  | 1.337E-06 | 4.774E-04 |
|---|
| kinase inhibitor activity | GO:0019210 |  | 1.655E-06 | 5.373E-04 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.719E-09 | 6.27E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.979E-09 | 7.257E-06 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 1.496E-08 | 1.819E-05 |
|---|
| kinase regulator activity | GO:0019207 |  | 3.067E-08 | 2.797E-05 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 9.036E-08 | 6.593E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.701E-07 | 1.034E-04 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 8.978E-07 | 4.679E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 2.457E-06 | 1.12E-03 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 3.224E-06 | 1.307E-03 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 6.436E-06 | 2.348E-03 |
|---|
| protein kinase inhibitor activity | GO:0004860 |  | 9.456E-06 | 3.136E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.414E-09 | 1.303E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.951E-07 | 2.347E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 2.972E-06 | 2.384E-03 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 4.708E-06 | 2.832E-03 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 8.393E-06 | 4.039E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 1.234E-05 | 4.95E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 1.85E-05 | 6.36E-03 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 1.85E-05 | 5.565E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.93E-05 | 5.16E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 2.589E-05 | 6.228E-03 |
|---|
| response to UV | GO:0009411 |  | 2.963E-05 | 6.48E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.614E-08 | 3.712E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.623E-07 | 5.317E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 6.378E-06 | 4.89E-03 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 9.334E-06 | 5.367E-03 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 1.451E-05 | 6.676E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 2.466E-05 | 9.455E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 3.696E-05 | 0.01214401 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 3.696E-05 | 0.01062601 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 3.696E-05 | 9.445E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.35E-05 | 0.01000551 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 5.169E-05 | 0.01080849 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.614E-08 | 3.712E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.623E-07 | 5.317E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 6.378E-06 | 4.89E-03 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 9.334E-06 | 5.367E-03 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 1.451E-05 | 6.676E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 2.466E-05 | 9.455E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 3.696E-05 | 0.01214401 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 3.696E-05 | 0.01062601 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 3.696E-05 | 9.445E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.35E-05 | 0.01000551 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 5.169E-05 | 0.01080849 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.614E-08 | 3.712E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.623E-07 | 5.317E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 6.378E-06 | 4.89E-03 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 9.334E-06 | 5.367E-03 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 1.451E-05 | 6.676E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 2.466E-05 | 9.455E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 3.696E-05 | 0.01214401 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 3.696E-05 | 0.01062601 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 3.696E-05 | 9.445E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.35E-05 | 0.01000551 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 5.169E-05 | 0.01080849 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.614E-08 | 3.712E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.623E-07 | 5.317E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 6.378E-06 | 4.89E-03 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 9.334E-06 | 5.367E-03 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 1.451E-05 | 6.676E-03 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 2.466E-05 | 9.455E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 3.696E-05 | 0.01214401 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 3.696E-05 | 0.01062601 |
|---|
| DNA damage response. signal transduction resulting in transcription | GO:0042772 |  | 3.696E-05 | 9.445E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.35E-05 | 0.01000551 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 5.169E-05 | 0.01080849 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.786E-08 | 4.237E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 5.186E-07 | 6.153E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 6.788E-06 | 5.369E-03 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 1.219E-05 | 7.232E-03 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 1.813E-05 | 8.602E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 2.345E-05 | 9.273E-03 |
|---|
| regulation of centrosome duplication | GO:0010824 |  | 2.345E-05 | 7.948E-03 |
|---|
| positive regulation of DNA repair | GO:0045739 |  | 3.513E-05 | 0.01042154 |
|---|
| regulation of centrosome cycle | GO:0046605 |  | 3.513E-05 | 9.264E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.349E-05 | 0.01032018 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.914E-05 | 0.01060071 |
|---|



