Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 2189-2-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1924 | 9.821e-03 | 1.174e-02 | 9.775e-02 |
|---|
| IPC-NIBC-129 | 0.2587 | 7.212e-02 | 9.372e-02 | 5.670e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2492 | 3.220e-02 | 4.409e-02 | 4.938e-01 |
|---|
| Loi_GPL570 | 0.1877 | 5.456e-02 | 5.416e-02 | 2.717e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1716 | 1.410e-04 | 6.690e-04 | 3.392e-03 |
|---|
| Parker_GPL1390 | 0.2315 | 1.093e-01 | 1.097e-01 | 5.641e-01 |
|---|
| Parker_GPL887 | 0.0550 | 8.209e-01 | 8.383e-01 | 8.735e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3933 | 1.378e-01 | 1.061e-01 | 6.222e-01 |
|---|
| Schmidt | 0.2007 | 9.553e-03 | 1.367e-02 | 1.262e-01 |
|---|
| Sotiriou | 0.3180 | 1.959e-03 | 5.320e-04 | 5.220e-02 |
|---|
| Wang | 0.1726 | 9.192e-02 | 6.232e-02 | 3.459e-01 |
|---|
| Zhang | 0.2397 | 1.164e-01 | 7.857e-02 | 2.746e-01 |
|---|
| Zhou | 0.4275 | 4.455e-02 | 4.565e-02 | 2.852e-01 |
|---|
Expression data for subnetwork 2189-2-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
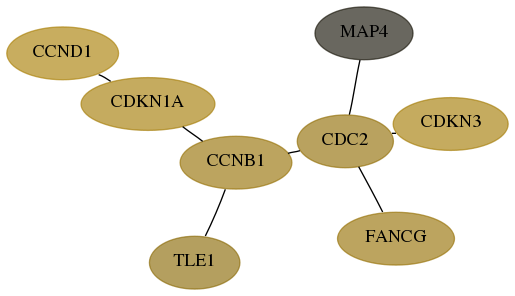
Score for each gene in subnetwork 2189-2-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| cdkn3 |   | 15 | 15 | 128 | 104 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.166 | -0.009 | 0.323 | 0.186 | 0.198 | 0.134 | 0.271 | 0.372 |
|---|
| map4 |   | 16 | 13 | 73 | 55 | 0.002 | 0.046 | 0.067 | -0.043 | 0.034 | 0.119 | 0.159 | -0.058 | -0.020 | 0.043 | 0.108 | -0.074 | 0.302 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| tle1 |   | 23 | 9 | 78 | 60 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | -0.028 | 0.068 | 0.025 |
|---|
| ccnb1 |   | 36 | 7 | 78 | 59 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | 0.152 | 0.167 | 0.398 |
|---|
| fancg |   | 1 | 164 | 358 | 358 | 0.081 | 0.214 | 0.140 | 0.118 | 0.100 | 0.152 | 0.239 | 0.215 | 0.173 | 0.118 | 0.162 | 0.070 | 0.344 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
GO Enrichment output for subnetwork 2189-2-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.821E-08 | 6.488E-05 |
|---|
| interphase | GO:0051325 |  | 3.169E-08 | 3.644E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.197E-07 | 9.177E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.411E-07 | 1.386E-04 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 6.041E-07 | 2.779E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.43E-06 | 5.481E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.43E-06 | 4.698E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.495E-06 | 4.3E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.901E-06 | 4.859E-04 |
|---|
| response to UV | GO:0009411 |  | 1.604E-05 | 3.689E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 4.898E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.308E-09 | 1.785E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.043E-08 | 3.718E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.329E-08 | 5.968E-05 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 3.476E-07 | 2.123E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 5.326E-07 | 2.602E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 6.997E-07 | 2.849E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.906E-07 | 2.759E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.11E-06 | 3.388E-04 |
|---|
| retrograde protein transport. ER to cytosol | GO:0030970 |  | 4.743E-06 | 1.288E-03 |
|---|
| response to UV | GO:0009411 |  | 7.871E-06 | 1.923E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.326E-05 | 2.946E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.138E-08 | 2.738E-05 |
|---|
| interphase | GO:0051325 |  | 1.332E-08 | 1.602E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.948E-08 | 3.968E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.191E-07 | 7.162E-05 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.272E-07 | 2.056E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 8.634E-07 | 3.462E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.011E-06 | 3.475E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.011E-06 | 3.04E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.438E-06 | 3.843E-04 |
|---|
| retrograde protein transport. ER to cytosol | GO:0030970 |  | 6.077E-06 | 1.462E-03 |
|---|
| response to UV | GO:0009411 |  | 1.015E-05 | 2.219E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.308E-09 | 1.785E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.043E-08 | 3.718E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.329E-08 | 5.968E-05 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 3.476E-07 | 2.123E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 5.326E-07 | 2.602E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 6.997E-07 | 2.849E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 7.906E-07 | 2.759E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.11E-06 | 3.388E-04 |
|---|
| retrograde protein transport. ER to cytosol | GO:0030970 |  | 4.743E-06 | 1.288E-03 |
|---|
| response to UV | GO:0009411 |  | 7.871E-06 | 1.923E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.326E-05 | 2.946E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.138E-08 | 2.738E-05 |
|---|
| interphase | GO:0051325 |  | 1.332E-08 | 1.602E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.948E-08 | 3.968E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.191E-07 | 7.162E-05 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.272E-07 | 2.056E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 8.634E-07 | 3.462E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.011E-06 | 3.475E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.011E-06 | 3.04E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.438E-06 | 3.843E-04 |
|---|
| retrograde protein transport. ER to cytosol | GO:0030970 |  | 6.077E-06 | 1.462E-03 |
|---|
| response to UV | GO:0009411 |  | 1.015E-05 | 2.219E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 9.823E-11 | 3.508E-07 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.89E-09 | 5.16E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 3.674E-09 | 4.373E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.076E-09 | 3.639E-06 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.082E-08 | 7.726E-06 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.453E-08 | 2.65E-05 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 6.342E-08 | 3.236E-05 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.699E-07 | 7.585E-05 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 2.14E-07 | 8.492E-05 |
|---|
| response to UV | GO:0009411 |  | 1.199E-06 | 4.28E-04 |
|---|
| retrograde protein transport. ER to cytosol | GO:0030970 |  | 1.933E-06 | 6.277E-04 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.418E-11 | 2.706E-07 |
|---|
| interphase | GO:0051325 |  | 8.403E-11 | 1.533E-07 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.678E-09 | 2.04E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 2.866E-09 | 2.614E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.392E-09 | 2.475E-06 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.861E-08 | 1.131E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.861E-08 | 1.491E-05 |
|---|
| response to UV | GO:0009411 |  | 7.208E-07 | 3.287E-04 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 1.597E-06 | 6.473E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.915E-06 | 6.984E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 2.21E-06 | 7.329E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.138E-08 | 2.738E-05 |
|---|
| interphase | GO:0051325 |  | 1.332E-08 | 1.602E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.948E-08 | 3.968E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.191E-07 | 7.162E-05 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 4.272E-07 | 2.056E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 8.634E-07 | 3.462E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.011E-06 | 3.475E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.011E-06 | 3.04E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.438E-06 | 3.843E-04 |
|---|
| retrograde protein transport. ER to cytosol | GO:0030970 |  | 6.077E-06 | 1.462E-03 |
|---|
| response to UV | GO:0009411 |  | 1.015E-05 | 2.219E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.821E-08 | 6.488E-05 |
|---|
| interphase | GO:0051325 |  | 3.169E-08 | 3.644E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.197E-07 | 9.177E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.411E-07 | 1.386E-04 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 6.041E-07 | 2.779E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.43E-06 | 5.481E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.43E-06 | 4.698E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.495E-06 | 4.3E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.901E-06 | 4.859E-04 |
|---|
| response to UV | GO:0009411 |  | 1.604E-05 | 3.689E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 4.898E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.821E-08 | 6.488E-05 |
|---|
| interphase | GO:0051325 |  | 3.169E-08 | 3.644E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.197E-07 | 9.177E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.411E-07 | 1.386E-04 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 6.041E-07 | 2.779E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.43E-06 | 5.481E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.43E-06 | 4.698E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.495E-06 | 4.3E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.901E-06 | 4.859E-04 |
|---|
| response to UV | GO:0009411 |  | 1.604E-05 | 3.689E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 4.898E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.821E-08 | 6.488E-05 |
|---|
| interphase | GO:0051325 |  | 3.169E-08 | 3.644E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.197E-07 | 9.177E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.411E-07 | 1.386E-04 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 6.041E-07 | 2.779E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.43E-06 | 5.481E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.43E-06 | 4.698E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.495E-06 | 4.3E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.901E-06 | 4.859E-04 |
|---|
| response to UV | GO:0009411 |  | 1.604E-05 | 3.689E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 4.898E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.821E-08 | 6.488E-05 |
|---|
| interphase | GO:0051325 |  | 3.169E-08 | 3.644E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.197E-07 | 9.177E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.411E-07 | 1.386E-04 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 6.041E-07 | 2.779E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.43E-06 | 5.481E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.43E-06 | 4.698E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.495E-06 | 4.3E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.901E-06 | 4.859E-04 |
|---|
| response to UV | GO:0009411 |  | 1.604E-05 | 3.689E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.343E-05 | 4.898E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.58E-08 | 3.749E-05 |
|---|
| interphase | GO:0051325 |  | 1.773E-08 | 2.103E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.882E-08 | 6.234E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.691E-07 | 1.003E-04 |
|---|
| cellular response to unfolded protein | GO:0034620 |  | 5.978E-07 | 2.837E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 9.725E-07 | 3.846E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.147E-06 | 3.888E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.477E-06 | 4.382E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.895E-06 | 4.998E-04 |
|---|
| response to UV | GO:0009411 |  | 1.167E-05 | 2.77E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.811E-05 | 3.906E-03 |
|---|



