Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1780-1-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1693 | 6.040e-03 | 7.192e-03 | 6.753e-02 |
|---|
| IPC-NIBC-129 | 0.2342 | 3.369e-02 | 4.174e-02 | 3.424e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2727 | 7.583e-03 | 1.200e-02 | 2.224e-01 |
|---|
| Loi_GPL570 | 0.2559 | 7.618e-02 | 7.569e-02 | 3.444e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1730 | 1.038e-03 | 3.804e-03 | 3.900e-02 |
|---|
| Parker_GPL1390 | 0.2868 | 1.501e-01 | 1.546e-01 | 6.504e-01 |
|---|
| Parker_GPL887 | 0.0153 | 9.662e-01 | 9.703e-01 | 9.856e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3851 | 1.500e-01 | 1.170e-01 | 6.502e-01 |
|---|
| Schmidt | 0.1705 | 1.119e-02 | 1.590e-02 | 1.425e-01 |
|---|
| Sotiriou | 0.3083 | 1.798e-03 | 4.770e-04 | 4.900e-02 |
|---|
| Wang | 0.2218 | 8.989e-02 | 6.072e-02 | 3.408e-01 |
|---|
| Zhang | 0.2313 | 5.726e-02 | 3.359e-02 | 1.445e-01 |
|---|
| Zhou | 0.4327 | 4.824e-02 | 4.983e-02 | 3.069e-01 |
|---|
Expression data for subnetwork 1780-1-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
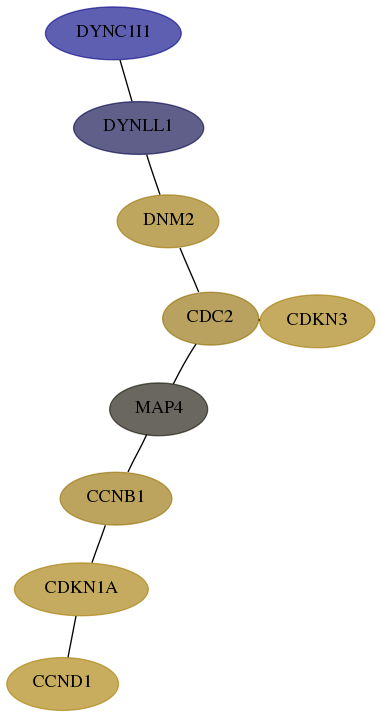
Score for each gene in subnetwork 1780-1-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| cdkn3 |   | 15 | 15 | 128 | 104 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.166 | -0.009 | 0.323 | 0.186 | 0.198 | 0.134 | 0.271 | 0.372 |
|---|
| map4 |   | 16 | 13 | 73 | 55 | 0.002 | 0.046 | 0.067 | -0.043 | 0.034 | 0.119 | 0.159 | -0.058 | -0.020 | 0.043 | 0.108 | -0.074 | 0.302 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| dync1i1 |   | 2 | 104 | 93 | 97 | -0.058 | 0.102 | 0.055 | 0.124 | 0.054 | 0.147 | -0.008 | 0.056 | 0.082 | 0.036 | 0.119 | 0.166 | -0.245 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| dnm2 |   | 6 | 43 | 93 | 81 | 0.089 | 0.127 | 0.114 | -0.049 | 0.065 | 0.251 | 0.291 | 0.177 | -0.039 | -0.048 | -0.099 | -0.044 | 0.262 |
|---|
| ccnb1 |   | 36 | 7 | 78 | 59 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | 0.152 | 0.167 | 0.398 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| dynll1 |   | 5 | 50 | 93 | 82 | -0.012 | 0.108 | 0.088 | 0.087 | 0.098 | 0.124 | 0.134 | 0.128 | 0.113 | 0.093 | 0.158 | 0.074 | 0.360 |
|---|
GO Enrichment output for subnetwork 1780-1-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.407E-10 | 3.235E-07 |
|---|
| interphase | GO:0051325 |  | 1.62E-10 | 1.863E-07 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.624E-09 | 2.779E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.376E-08 | 4.241E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.487E-07 | 6.838E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.336E-06 | 5.123E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.734E-06 | 5.696E-04 |
|---|
| response to UV | GO:0009411 |  | 1.13E-05 | 3.248E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.864E-05 | 4.763E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.484E-05 | 5.712E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 2.484E-05 | 5.193E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.862E-11 | 1.188E-07 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.003E-09 | 2.446E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.559E-08 | 2.084E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.164E-08 | 3.764E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 5.423E-07 | 2.65E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 6.961E-07 | 2.834E-04 |
|---|
| response to UV | GO:0009411 |  | 6.934E-06 | 2.42E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.22E-05 | 3.727E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.317E-05 | 3.575E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.376E-05 | 3.362E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.564E-05 | 3.474E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.011E-11 | 1.446E-07 |
|---|
| interphase | GO:0051325 |  | 7.27E-11 | 8.746E-08 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.673E-09 | 2.144E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.414E-08 | 2.053E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.22E-08 | 3.955E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 6.779E-07 | 2.718E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 8.7E-07 | 2.99E-04 |
|---|
| response to UV | GO:0009411 |  | 7.739E-06 | 2.328E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.422E-05 | 3.803E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.644E-05 | 3.955E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.718E-05 | 3.758E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.862E-11 | 1.188E-07 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.003E-09 | 2.446E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.559E-08 | 2.084E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.164E-08 | 3.764E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 5.423E-07 | 2.65E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 6.961E-07 | 2.834E-04 |
|---|
| response to UV | GO:0009411 |  | 6.934E-06 | 2.42E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.22E-05 | 3.727E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.317E-05 | 3.575E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.376E-05 | 3.362E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.564E-05 | 3.474E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.011E-11 | 1.446E-07 |
|---|
| interphase | GO:0051325 |  | 7.27E-11 | 8.746E-08 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.673E-09 | 2.144E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.414E-08 | 2.053E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.22E-08 | 3.955E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 6.779E-07 | 2.718E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 8.7E-07 | 2.99E-04 |
|---|
| response to UV | GO:0009411 |  | 7.739E-06 | 2.328E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.422E-05 | 3.803E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.644E-05 | 3.955E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.718E-05 | 3.758E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.846E-13 | 2.445E-09 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 8.464E-11 | 1.511E-07 |
|---|
| cytoplasmic dynein complex | GO:0005868 |  | 1.05E-09 | 1.25E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.767E-09 | 5.148E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 5.876E-09 | 4.197E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.131E-09 | 4.839E-06 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.73E-08 | 8.826E-06 |
|---|
| microtubule associated complex | GO:0005875 |  | 1.211E-07 | 5.405E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.61E-07 | 6.389E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.851E-07 | 6.611E-05 |
|---|
| dynein complex | GO:0030286 |  | 3.42E-07 | 1.11E-04 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.126E-12 | 4.106E-09 |
|---|
| interphase | GO:0051325 |  | 1.309E-12 | 2.387E-09 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 7.449E-11 | 9.058E-08 |
|---|
| cytoplasmic dynein complex | GO:0005868 |  | 2.194E-09 | 2.001E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.016E-09 | 3.66E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 6.139E-09 | 3.732E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.014E-08 | 5.282E-06 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 3.983E-08 | 1.816E-05 |
|---|
| microtubule associated complex | GO:0005875 |  | 1.389E-07 | 5.629E-05 |
|---|
| dynein complex | GO:0030286 |  | 2.509E-07 | 9.153E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 3.571E-07 | 1.184E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.011E-11 | 1.446E-07 |
|---|
| interphase | GO:0051325 |  | 7.27E-11 | 8.746E-08 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.673E-09 | 2.144E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.414E-08 | 2.053E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.22E-08 | 3.955E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 6.779E-07 | 2.718E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 8.7E-07 | 2.99E-04 |
|---|
| response to UV | GO:0009411 |  | 7.739E-06 | 2.328E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.422E-05 | 3.803E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.644E-05 | 3.955E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.718E-05 | 3.758E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.407E-10 | 3.235E-07 |
|---|
| interphase | GO:0051325 |  | 1.62E-10 | 1.863E-07 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.624E-09 | 2.779E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.376E-08 | 4.241E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.487E-07 | 6.838E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.336E-06 | 5.123E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.734E-06 | 5.696E-04 |
|---|
| response to UV | GO:0009411 |  | 1.13E-05 | 3.248E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.864E-05 | 4.763E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.484E-05 | 5.712E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 2.484E-05 | 5.193E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.407E-10 | 3.235E-07 |
|---|
| interphase | GO:0051325 |  | 1.62E-10 | 1.863E-07 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.624E-09 | 2.779E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.376E-08 | 4.241E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.487E-07 | 6.838E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.336E-06 | 5.123E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.734E-06 | 5.696E-04 |
|---|
| response to UV | GO:0009411 |  | 1.13E-05 | 3.248E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.864E-05 | 4.763E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.484E-05 | 5.712E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 2.484E-05 | 5.193E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.407E-10 | 3.235E-07 |
|---|
| interphase | GO:0051325 |  | 1.62E-10 | 1.863E-07 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.624E-09 | 2.779E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.376E-08 | 4.241E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.487E-07 | 6.838E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.336E-06 | 5.123E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.734E-06 | 5.696E-04 |
|---|
| response to UV | GO:0009411 |  | 1.13E-05 | 3.248E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.864E-05 | 4.763E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.484E-05 | 5.712E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 2.484E-05 | 5.193E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.407E-10 | 3.235E-07 |
|---|
| interphase | GO:0051325 |  | 1.62E-10 | 1.863E-07 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.624E-09 | 2.779E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.376E-08 | 4.241E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.487E-07 | 6.838E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.336E-06 | 5.123E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.734E-06 | 5.696E-04 |
|---|
| response to UV | GO:0009411 |  | 1.13E-05 | 3.248E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.864E-05 | 4.763E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.484E-05 | 5.712E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 2.484E-05 | 5.193E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.003E-11 | 1.662E-07 |
|---|
| interphase | GO:0051325 |  | 8.051E-11 | 9.552E-08 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.166E-09 | 1.714E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.855E-08 | 2.88E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.042E-07 | 4.947E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.179E-06 | 4.664E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.498E-06 | 5.079E-04 |
|---|
| response to UV | GO:0009411 |  | 8.218E-06 | 2.438E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.441E-05 | 3.798E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 1.92E-05 | 4.556E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.296E-05 | 4.953E-03 |
|---|



